Yeast-Based Fluorescent Sensors for the Simultaneous Detection of Estrogenic and Androgenic Compounds, Coupled with High-Performance Thin Layer Chromatography
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Yeast Strains, Plasmids, and Growth Conditions
2.3. Endocrine Assay in 96-Well Plates
2.4. Calculation of the Corrected Fluorescence and the Reporter Gene Induction in 96-Well Plates
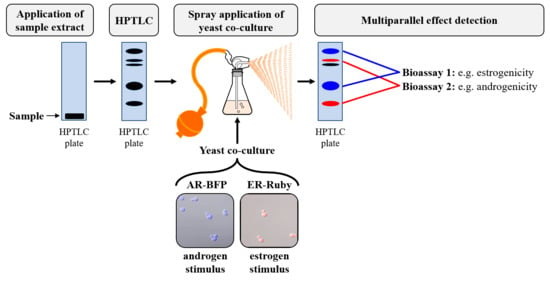
2.5. Endocrine Assay on the HPTLC Plate Surface
2.6. Preparation of Wastewater Samples
2.7. Fluorescent Microscopy
2.8. Data Processing and Statistical Analysis
3. Results
3.1. Sensor Strain Characterization in a 96-Well Plate Assay
3.2. Simultaneous Detection of Estrogenic and Androgenic Activities—Model Compounds
3.3. Simultaneous Detection of Estrogenic and Androgenic Activities—Wastewater Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Da Silva, A.P.A.; De Oliveira, C.D.L.; Quirino, A.M.S.; da Silva, F.D.M.; de Aquino Saraiva, R.D.; Silva-Cavalcanti, J.S. Endocrine disruptors in aquatic environment: Effects and consequences on the biodiversity of fish and amphibian species. Aquat. Sci. Technol. 2018, 6, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Kerdivel, G.; Habauzit, D.; Pakdel, F. Assessment and molecular actions of endocrine-disrupting chemicals that interfere with estrogen receptor pathways. Int. J. Endocrinol. 2013, 2013, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, A.M.; Sonnenschein, C. Environmental causes of cancer: Endocrine disruptors as carcinogens. Nat. Rev. Endocrinol. 2010, 6, 363–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R., Jr.; Lee, D.H.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; Welshons, W.V.; et al. Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012, 33, 378–455. [Google Scholar] [CrossRef] [PubMed]
- Colborn, T.; Saal, F.S.V.; Soto, A.M. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ. Health Perspect. 1993, 101, 378–384. [Google Scholar] [CrossRef]
- Casals-Casas, C.; Desvergne, B. Endocrine disruptors: From endocrine to metabolic disruption. Annu. Rev. Physiol. 2011, 73, 135–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arlos, M.J.; Parker, W.J.; Bicudo, J.R.; Law, P.; Hicks, K.A.; Fuzzen, M.L.; Andrews, S.A.; Servos, M.R. Modeling the exposure of wild fish to endocrine active chemicals: Potential linkages of total estrogenicity to field-observed intersex. Water Res. 2018, 139, 187–197. [Google Scholar] [CrossRef]
- Svenson, A.; Allard, A.S.; Ek, M. Removal of estrogenicity in Swedish municipal sewage treatment plants. Water Res. 2003, 37, 4433–4443. [Google Scholar] [CrossRef]
- Sumpter, J.P.; Johnson, A.C. 10th Anniversary Perspective: Reflections on endocrine disruption in the aquatic environment: From known knowns to unknown unknowns (and many things in between). J. Environ. Monit. 2008, 10, 1476–1485. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen receptor structure, function and biology: From bench to bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar] [PubMed]
- Yaşar, P.; Ayaz, G.; User, S.D.; Güpür, G.; Muyan, M. Molecular mechanism of estrogen-estrogen receptor signaling. Reprod. Med. Biol. 2017, 16, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Campana, C.; Pezzi, V.; Rainey, W.E. Cell-based assays for screening androgen receptor ligands. Semin. Reprod. Med. 2015, 33, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riegraf, C.; Reifferscheid, G.; Belkin, S.; Moscovici, L.; Shakibai, D.; Hollert, H.; Buchinger, S. Combination of yeast-based in vitro screens with high-performance thin-layer chromatography as a novel tool for the detection of hormonal and dioxin-like compounds. Anal. Chim. Acta 2019, 1081, 218–230. [Google Scholar] [CrossRef]
- Adeniran, A.; Sherer, M.; Tyo, K.E.J. Yeast-based biosensors: Design and applications. FEMS Yeast Res. 2015, 15, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, A.J.; Simon, E.; Schifferli, A.; Schönborn, A.; Vermeirssen, E.L.M. Estrogenic activity of food contact materials—Evaluation of 20 chemicals using a yeast estrogen screen on HPTLC or 96-well plates. Anal. Bioanal. Chem. 2020, 412, 4527–4536. [Google Scholar] [CrossRef]
- Shakibai, D.; Riegraf, C.; Moscovici, L.; Reifferscheid, G.; Buchinger, S.; Belkin, S. Coupling high-performance thin-layer chromatography with bacterial genotoxicity bioreporters. Environ. Sci. Technol. 2019, 53, 6410–6419. [Google Scholar] [CrossRef] [PubMed]
- Klingelhöfer, I.; Morlock, G.E. Sharp-bounded zones link to the effect in planar chromatography-bioassay-mass spectrometry. J. Chromatogr. A 2014, 1360, 288–295. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Morlock, G.E. Bioprofiling of surface/wastewater and bioquantitation of discovered endocrine-active compounds by streamlined direct bioautography. Anal. Chem. 2015, 87, 11098–11104. [Google Scholar] [CrossRef] [PubMed]
- Chamas, A.; Pham, H.T.M.; Jähne, M.; Hettwer, K.; Gehrmann, L.; Tuerk, J.; Uhlig, S.; Simon, K.; Baronian, K.; Kunze, G. Separation and identification of hormone-active compounds using a combination of chromatographic separation and yeast-based reporter assay. Sci. Total. Environ. 2017, 605, 507–513. [Google Scholar] [CrossRef]
- Schoenborn, A.; Schmid, P.; Bräm, S.; Reifferscheid, G.; Ohlig, M.; Buchinger, S. Unprecedented sensitivity of the planar yeast estrogen screen by using a spray-on technology. J. Chromatogr. A 2017, 1530, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, J.; Gupta, R.K.; Layton, A.C.; Patterson, S.S.; Ripp, S.A.; Saidak, L.; Simpson, M.L.; Schultz, T.W.; Sayler, G.S. Use of Saccharomyces cerevisiae BLYES Expressing Bacterial Bioluminescence for Rapid, Sensitive Detection of Estrogenic Compounds. Appl. Environ. Microbiol. 2005, 71, 4455–4460. [Google Scholar] [CrossRef] [Green Version]
- Eldridge, M.L.; Sanseverino, J.; Layton, A.C.; Easter, J.P.; Schultz, T.W.; Sayler, G.S. Saccharomyces cerevisiae BLYAS, a new bioluminescent bioreporter for detection of androgenic compounds. Appl. Environ. Microbiol. 2007, 73, 6012–6018. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lim, W.A.; Thorn, K.S. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS ONE 2013, 8, e67902. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; Volume 1. [Google Scholar]
- Routledge, E.J.; Sumpter, J.P. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ. Toxicol. Chem. 1996, 15, 241–248. [Google Scholar] [CrossRef]
- Purvis, I.J.; Chotai, D.; Dykes, C.W.; Lubahn, D.B.; French, F.S.; Wilson, E.M.; Hobden, A.N. An androgen-inducible expression system for Saccharomyces cerevisiae. Gene 1991, 106, 35–42. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef]
- Dymond, J.S. Saccharomyces cerevisiae growth media. Methods Enzymol. 2013, 533, 191–204. [Google Scholar] [CrossRef]
- Spira, D.; Reifferscheid, G.; Buchinger, S. Combination of high-performance thin-layer chromatography with a specific bioassay—A tool for effect-directed analysis. J. Planar Chromatogr. Mod. TLC 2013, 26, 395–401. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 11 May 2020).
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org/authors.html (accessed on 11 May 2020).
- Buchinger, S.; Spira, D.; Bröder, K.; Schlüsener, M.; Ternes, T.; Reifferscheid, G. Direct coupling of thin-layer chromatography with a bioassay for the detection of estrogenic compounds: Applications for effect-directed analysis. Anal. Chem. 2013, 85, 7248–7256. [Google Scholar] [CrossRef]
- Dausend, C.; Weins, C.; Frimmel, F. A New bioautographic screening method for the detection of estrogenic compounds. Chromatographia 2004, 60, 207–211. [Google Scholar] [CrossRef]
- Jarosova, B.; Érseková, A.; Hilscherová, K.; Loos, R.; Gawlik, B.M.; Giesy, J.P.; Bláha, L. Europe-wide survey of estrogenicity in wastewater treatment plant effluents: The need for the effect-based monitoring. Environ. Sci. Pollut. Res. 2014, 21, 10970–10982. [Google Scholar] [CrossRef]
- Leusch, F.D.; Chapman, H.F.; Heuvel, M.R.V.D.; Tan, B.L.; Gooneratne, S.R.; Tremblay, L.A. Bioassay-derived androgenic and estrogenic activity in municipal sewage in Australia and New Zealand. Ecotoxicol. Environ. Saf. 2006, 65, 403–411. [Google Scholar] [CrossRef]
- Coldham, N.G.; Dave, M.; Sivapathasundaram, S.; McDonnell, D.P.; Connor, C.; Sauer, M.J. Evaluation of a recombinant yeast cell estrogen screening assay. Environ. Health Perspect. 1997, 105, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Miller III, C.A.; Tan, X.; Wilson, M.; Bhattacharyya, S.; Ludwig, S. Single plasmids expressing human steroid hormone receptors and a reporter gene for use in yeast signaling assays. Plasmid 2010, 63, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Bovee, T.F.; Helsdingen, R.J.R.; Hamers, A.R.M.; Van Duursen, M.B.M.; Nielen, M.W.F.; Hoogenboom, R.L.A.P. A new highly specific and robust yeast androgen bioassay for the detection of agonists and antagonists. Anal. Bioanal. Chem. 2007, 389, 1549–1558. [Google Scholar] [CrossRef] [Green Version]
- Bovee, T.F.; Helsdingen, R.J.; Koks, P.D.; Kuiper, H.A.; Hoogenboom, R.L.; Keijer, J. Development of a rapid yeast estrogen bioassay, based on the expression of green fluorescent protein. Gene 2004, 325, 187–200. [Google Scholar] [CrossRef]
- Handa, R.J.; Pak, T.R.; Kudwa, A.E.; Lund, T.D.; Hinds, L. An alternate pathway for androgen regulation of brain function: Activation of estrogen receptor beta by the metabolite of dihydrotestosterone, 5α-androstane-3β,17β-diol. Horm. Behav. 2008, 53, 741–752. [Google Scholar] [CrossRef] [Green Version]
- Beresford, N.; Routledge, E.J.; Harris, C.A.; Sumpter, J.P. Issues arising when interpreting results from an in vitro assay for estrogenic activity. Toxicol. Appl. Pharmacol. 2000, 162, 22–33. [Google Scholar] [CrossRef] [PubMed]









| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| S. cerevisiae parental strains | ||
| hER | MATa; leu2; his3; Human estrogen receptor gene in the chromosome. | [25] |
| hAR | BJ 1991 MATa; prb1-1122; pep4-3; leu2; trp1; ura3-52; GAL Human estrogen receptor gene in the chromosome. | [26] |
| Parental plasmids | ||
| pFA6a-link-yomRuby2/yomTagBFP2/yoEGFP | containing Ruby, BFP, EGFP gene respectively | Addgene #44858 #44839#44836 respectively [23] |
| pUTK407 | Contains the luxA and luxB genes expressed from the bidirectional promoters GPD and ADH1 separated with two EREs. | [21] |
| pUTK420 | Contains the luxA and luxB genes expressed from the bidirectional promoters GPD and ADH1 separated with four AREs. | [22] |
| ER and AR reporter plasmids | ||
| ER fluorescent reporter (FP) | pUTK407 in which luxB was substituted by EGFP/Ruby/BFP gene. | This study |
| AR fluorescent reporter (FP) | pUTK420 in which luxB was substituted by EGFP/Ruby/BFP gene. | This study |
| Fluorescent sensor strains | ||
| hER-EGFP | Contains the EGFP gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| hER-Ruby | Contains the Ruby gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| hER-BFP | Contains the BFP gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| hAR-EGFP | Contains the EGFP gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| hAR-Ruby | Contains the Ruby gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| hAR-BFP | Contains the BFP gene contred by ADH1 promoter and the luxA gene expressed from GPD promoter, with two repeats of EREs. | This study |
| Single Strain | Both Strains | Rf Values | ||||
|---|---|---|---|---|---|---|
| Individual | Mix | Individual | Mix | |||
| E3 | Mean LOD (ng) | 0.82 | 1.43 | 1.7 | 3.6 | 0.19 |
| SE (ng) | 0.02 | 0.09 | 0.4 | 1.9 | ||
| E2 | Mean LOD (ng) | 0.0081 | 0.012 | 0.03 | 0.04 | 0.60 |
| SE (ng) | 0.0009 | 0.001 | 0.02 | 0.02 | ||
| E1 | Mean LOD (ng) | 0.0120 | 0.016 | 0.05 | 0.06 | 0.76 |
| SE (ng) | 0.0002 | 0.002 | 0.03 | 0.04 | ||
| Mean LOQ (ng) | 0.042 | 0.058 | ||||
| SE (ng) | 0.003 | 0.008 | ||||
| Testosterone | Mean LOD (ng) | 0.3 | 0.70 | 0.8 (*) | 0.9 | 0.46 |
| SE (ng) | 0.2 | 0.04 | 0.2 | 0.2 | ||
| DHT | Mean LOD (ng) | <0.5 | <0.5 | 0.3 (*) | 0.4 | 0.61 |
| SE (ng) | 0.1 | 0.1 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moscovici, L.; Riegraf, C.; Abu-Rmailah, N.; Atias, H.; Shakibai, D.; Buchinger, S.; Reifferscheid, G.; Belkin, S. Yeast-Based Fluorescent Sensors for the Simultaneous Detection of Estrogenic and Androgenic Compounds, Coupled with High-Performance Thin Layer Chromatography. Biosensors 2020, 10, 169. https://doi.org/10.3390/bios10110169
Moscovici L, Riegraf C, Abu-Rmailah N, Atias H, Shakibai D, Buchinger S, Reifferscheid G, Belkin S. Yeast-Based Fluorescent Sensors for the Simultaneous Detection of Estrogenic and Androgenic Compounds, Coupled with High-Performance Thin Layer Chromatography. Biosensors. 2020; 10(11):169. https://doi.org/10.3390/bios10110169
Chicago/Turabian StyleMoscovici, Liat, Carolin Riegraf, Nidaa Abu-Rmailah, Hadas Atias, Dror Shakibai, Sebastian Buchinger, Georg Reifferscheid, and Shimshon Belkin. 2020. "Yeast-Based Fluorescent Sensors for the Simultaneous Detection of Estrogenic and Androgenic Compounds, Coupled with High-Performance Thin Layer Chromatography" Biosensors 10, no. 11: 169. https://doi.org/10.3390/bios10110169
APA StyleMoscovici, L., Riegraf, C., Abu-Rmailah, N., Atias, H., Shakibai, D., Buchinger, S., Reifferscheid, G., & Belkin, S. (2020). Yeast-Based Fluorescent Sensors for the Simultaneous Detection of Estrogenic and Androgenic Compounds, Coupled with High-Performance Thin Layer Chromatography. Biosensors, 10(11), 169. https://doi.org/10.3390/bios10110169