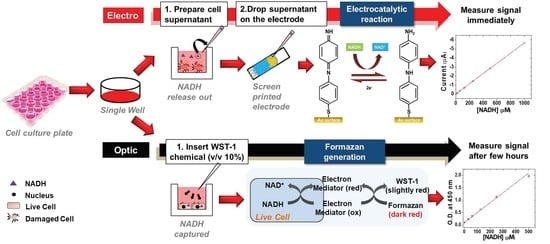
Non-Destructive Monitoring via Electrochemical NADH Detection in Murine Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Apparatus and Electrode
2.3. Surface Modification of the SPE
2.4. In Vitro Studies
2.4.1. Cell Culture and WST-1 Viability Assay
2.4.2. Quantification of NADH in Cell Supernatants
2.5. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Victor, V.M.; Apostolova, N.; Herance, R.; Hernandez-Mijares, A.; Rocha, M. Oxidative stress and mitochondrial dysfunction in atherosclerosis: Mitochondria-targeted antioxidants as potential therapy. Curr. Med. Chem. 2009, 16, 4654–4667. [Google Scholar] [CrossRef] [PubMed]
- Limongelli, G.; Masarone, D.; D’Alessandro, R.; Elliott, P.M. Mitochondrial diseases and the heart: An overview of molecular basis, diagnosis, treatment and clinical course. Future Cardiol. 2012, 8, 71–88. [Google Scholar] [CrossRef]
- Ma, Z.A.; Zhao, Z.; Turk, J. Mitochondrial dysfunction and β-cell failure in type 2 diabetes mellitus. Exp. Diabetes Res. 2011, 2012, 703538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, A.M.; Joanisse, D.R.; Baillot, R.G.; Hood, D.A. Mitochondrial dysregulation in the pathogenesis of diabetes: Potential for mitochondrial biogenesis-mediated interventions. Exp. Diabetes Res. 2011, 2012, 642038. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L. Metabolic syndrome and mitochondrial function: Molecular replacement and antioxidant supplements to prevent membrane peroxidation and restore mitochondrial function. J. Cell. Biochem. 2007, 100, 1352–1369. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Kim, D.I.; Kim, S.H.; Lee, H.; Lee, K.S.; Cho, S.H.; Lee, Y.C. NLRP3 inflammasome activation by mitochondrial ROS in bronchial epithelial cells is required for allergic inflammation. Cell Death Dis. 2014, 5, e1498. [Google Scholar] [CrossRef] [Green Version]
- Marshall, J.D.; Bazan, I.; Zhang, Y.; Fares, W.H.; Lee, P.J. Mitochondrial dysfunction and pulmonary hypertension: Cause, effect, or both. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 314, L782–L796. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, T.; Baur, J.A.; Perez, E.; Matsui, T.; Carmona, J.J.; Lamming, D.W.; Souza-Pinto, N.C.; Bohr, V.A.; Rosenzweig, A.; et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 2007, 130, 1095–1107. [Google Scholar] [CrossRef] [Green Version]
- Adriouch, S.; Hubert, S.; Pechberty, S.; Koch-Nolte, F.; Haag, F.; Seman, M. NAD+ released during inflammation participates in T cell homeostasis by inducing ART2-mediated death of naive T cells in vivo. J. Immunol. 2007, 179, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Stienen, G.J.; Kiers, J.L.; Bottinelli, R.; Reggiani, C. Myofibrillar ATPase activity in skinned human skeletal muscle fibres: Fibre type and temperature dependence. J. Physiol. 1996, 493, 299–307. [Google Scholar] [CrossRef]
- Barlow, C.H.; Chance, B. Ischemic areas in perfused rat hearts: Measurement by NADH fluorescence photography. Science 1976, 193, 909–910. [Google Scholar] [CrossRef] [PubMed]
- Young, I.G.; Jaworowski, A.; Poulis, M.I. Amplification of the respiratory NADH dehydrogenase of Escherichia coli by gene cloning. Gene 1978, 4, 25–36. [Google Scholar] [CrossRef]
- Raj, C.R.; Ohsaka, T. Electrocatalytic sensing of NADH at an in situ functionalized self-assembled monolayer on gold electrode. Electrochem. Commun. 2001, 3, 633–638. [Google Scholar]
- Mayevsky, A.; Rogatsky, G.G. Mitochondrial function in vivo evaluated by NADH fluorescence: From animal models to human studies. Am. J. Physiol. Cell Physiol. 2007, 292, C615–C640. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Barman, K.; Jasimuddin, S. Electrochemical detection of adenine and guanine using a self-assembled copper (II)–thiophenyl-azo-imidazole complex monolayer modified gold electrode. RSC Adv. 2014, 4, 49819–49826. [Google Scholar] [CrossRef]
- Sánchez-Tirado, E.; Salvo, C.; González-Cortés, A.; Yáñez-Sedeño, P.; Langa, F.; Pingarrón, J.M. Electrochemical immunosensor for simultaneous determination of interleukin-1 beta and tumor necrosis factor alpha in serum and saliva using dual screen printed electrodes modified with functionalized double–walled carbon nanotubes. Anal. Chim. Acta 2017, 959, 66–73. [Google Scholar] [CrossRef]
- Sprules, S.D.; Hart, J.P.; Pittson, R.; Wring, S.A. Evaluation of a new disposable screen-printed sensor strip for the measurement of NADH and its modification to produce a lactate biosensor employing microliter volumes. Electroanalysis 1996, 8, 539–543. [Google Scholar] [CrossRef]
- Yamada, K.; Hara, N.; Shibata, T.; Osago, H.; Tsuchiya, M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal. Biochem. 2006, 352, 282–285. [Google Scholar] [CrossRef]
- Song, J.; Kim, W.; Kim, Y.B.; Kim, B.; Lee, K. Time course of polyhexamethyleneguanidine phosphate-induced lung inflammation and fibrosis in mice. Toxicol. Appl. Pharmacol. 2018, 345, 94–102. [Google Scholar] [CrossRef]
- Jain, P.; Chakma, B.; Patra, S.; Goswami, P. Hairpin stabilized fluorescent silver nanoclusters for quantitative detection of NAD+ and monitoring NAD+/NADH based enzymatic reactions. Anal. Chim. Acta 2017, 956, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Tığ, G.A. Highly sensitive amperometric biosensor for determination of NADH and ethanol based on Au-Ag nanoparticles/poly(L-Cysteine)/reduced graphene oxide nanocomposite. Talanta 2017, 175, 382–389. [Google Scholar]
- Selvarani, K.; Prabhakaran, A.; Arumugam, P.; Berchmans, S.; Nayak, P. 2D MoSe2 sheets embedded over a high surface graphene hybrid for the amperometric detection of NADH. Mikrochim. Acta 2018, 185, 411. [Google Scholar] [CrossRef] [PubMed]
- Vukojević, V.; Djurdjić, S.; Ognjanović, M.; Antić, B.; Kalcher, K.; Mutić, J.; Stanković, D.M. RuO2/graphene nanoribbon composite supported on screen printed electrode with enhanced electrocatalytic performances toward ethanol and NADH biosensing. Biosens. Bioelectron. 2018, 117, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Turner, A.P.; Mak, W.C. Positively-charged hierarchical PEDOT interface with enhanced electrode kinetics for NADH-based biosensors. Biosens. Bioelectron. 2018, 120, 115–121. [Google Scholar] [CrossRef]
- Chamchoy, K.; Pakotiprapha, D.; Pumirat, P.; Leartsakulpanich, U. Application of WST-8 based colorimetric NAD (P) H detection for quantitative dehydrogenase assays. BMC Biochem. 2019, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- You, S.H.; Lim, H.D.; Cheong, D.E.; Kim, E.S.; Kim, G.J. Rapid and sensitive detection of NADPH via mBFP-mediated enhancement of its fluorescence. PLoS ONE 2019, 14, e0212061. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Wang, Y.; Du, N.; Jiang, D.; Ge, Q.; Wu, M.; Yu, H.; Xu, B. An electricalchemical method to detect the branch-chain aminotransferases activity in lactic acid bacteria. Food Chem. 2019, 297, 125035. [Google Scholar] [CrossRef]
- Wilkening, S.; Schmitt, F.-J.; Lenz, O.; Zebger, I.; Horch, M.; Friedrich, T. Discriminating changes in intracellular NADH/NAD+ levels due to anoxicity and H2 supply in R. eutropha cells using the Frex fluorescence sensor. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 148062. [Google Scholar] [CrossRef]
- Li, X.; Kan, X. A boronic acid carbon nanodots/poly(thionine) sensing platform for the accurate and reliable detection of NADH. Bioelectrochemistrym 2019, 130, 107344. [Google Scholar] [CrossRef]
- Thiruppathi, M.; Lin, P.Y.; Chou, Y.T.; Ho, H.Y.; Wu, L.C.; Ho, J.A. Simple aminophenol-based electrochemical probes for non-enzymatic, dual amperometric detection of NADH and hydrogen peroxide. Talanta 2019, 200, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, X.; Yin, C.; Li, W.; Qin, X.; Chen, C. A dual-signal output ratiometric electrochemiluminescent sensor for NADH detection. Analyst 2019, 144, 5215–5222. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hou, J.; Wang, Z.; Zhou, Q.; Xu, C. Porous PtAg nanoshells/reduced graphene oxide based biosensors for low-potential detection of NADH. Mikrochim. Acta 2020, 187, 544. [Google Scholar] [CrossRef] [PubMed]
- Titoiu, A.M.; Necula-Petrareanu, G.; Visinescu, D.; Dinca, V.; Bonciu, A.; Mihailescu, C.N.; Purcarea, C.; Boukherroub, R.; Szunerits, S.; Vasilescu, A. Flow injection enzymatic biosensor for aldehydes based on a Meldola Blue-Ni complex electrochemical mediator. Mikrochim. Acta 2020, 187, 550. [Google Scholar] [CrossRef]
- Liu, B.W.; Huang, P.; Wu, F.Y. Rapid visual detection for nitroreductase based on the copper ions-induced and NADH-mediated aggregation of gold-silver alloy nanoparticles. Talanta 2021, 234, 122681. [Google Scholar] [CrossRef]
- Liu, B.W.; Huang, P.C.; Wu, F.Y. A novel light-controlled colorimetric detection assay for nitroreductase based on p-aminophenol-catalyzed and NADH-mediated synthesis of silver nanoparticles. Anal. Methods 2021, 13, 2223–2228. [Google Scholar] [CrossRef]
- Moshirian-Farahi, S.S.; Zamani, H.A.; Abedi, M.R. Highly sensitive voltammetric determination of NADH based on N-CQDs decorated SnO2/ionic liquid/carbon paste electrode. Nanotechnology 2021. [Google Scholar] [CrossRef]
- Wang, C.; Wang, T.; Li, Z.; Xu, X.; Zhang, X.; Li, D. An Electrochemical Enzyme Biosensor for Ammonium Detection in Aquaculture Using Screen-Printed Electrode Modified by Gold Nanoparticle/Polymethylene Blue. Biosensors 2021, 11, 335. [Google Scholar] [CrossRef]





| Year | Type | Specific Methods/Material | LOD | Ref. |
|---|---|---|---|---|
| 2017 | Electrochemical | Silver nanocluster | 22.3 μM | [21] |
| Electrochemical | Au–Ag nanoparticles/poly L-cysteine/reduced graphene oxide nanocomposite | 1.05 mM | [22] | |
| 2018 | Electrochemical | Graphical abstract Schematic of the 2D MoSe2/HEG | 1 uM | [23] |
| Electrochemical | RuO2-GNR/SPCE | 0.52 μM | [24] | |
| Electrochemical | Hierarchically structured PEDOT CMs electrodes | 5.3 μM | [25] | |
| 2019 | Optical | WST-8 and UV spectrophotometric methods | 0.32, 1.65 nM | [26] |
| Optical | ELISA kit + mBFP protein | 2 pM | [27] | |
| Electrochemical | MWCNTs–CS/GCE electrode | 0.12 μM | [28] | |
| Electrochemical | Frex fluorescence sensor | 100 μM | [29] | |
| Electrochemical | boronic acid functionalized carbon nanodots and poly(thionine) on an electrode surface | 0.15 μM | [30] | |
| Electrochemical | pre-anodized screen-printed carbon electrode | 28.9 μM | [31] | |
| Electrochemical | a dual-signal-output ratiometric ECL sensor | 2.5 μM | [32] | |
| 2020 | Electrochemical | PtAg nanoshells supported on reduced graphene oxide (PtAg/rGO) | 0.2 μM | [33] |
| Electrochemical | screen-printed CNF electrode | 0.5 μM | [34] | |
| 2021 | Optical | Au80Ag20 NPs | 0.23 μg/mL | [35] |
| Optical | novel light-controlled colorimetric detection assay | 0.27 μg/mL | [36] | |
| Electrochemical | N-CQDs decorated SnO2/ionic liquid/carbon paste electrode | 0.8 nM | [37] | |
| Electrochemical | SPCE/AuNPs/PMB | 0.4 mM | [38] | |
| 2022 | Electrochemical | SPE/NPQD/double-polymerized electrocatalytic assay | 0.45 μM | The present work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.K.; Suh, H.N.; Yoon, S.H.; Lee, K.H.; Ahn, S.Y.; Kim, H.J.; Kim, S.H. Non-Destructive Monitoring via Electrochemical NADH Detection in Murine Cells. Biosensors 2022, 12, 107. https://doi.org/10.3390/bios12020107
Lee JK, Suh HN, Yoon SH, Lee KH, Ahn SY, Kim HJ, Kim SH. Non-Destructive Monitoring via Electrochemical NADH Detection in Murine Cells. Biosensors. 2022; 12(2):107. https://doi.org/10.3390/bios12020107
Chicago/Turabian StyleLee, Ju Kyung, Han Na Suh, Sung Hoon Yoon, Kyu Hong Lee, Sae Young Ahn, Hyung Jin Kim, and Sang Hee Kim. 2022. "Non-Destructive Monitoring via Electrochemical NADH Detection in Murine Cells" Biosensors 12, no. 2: 107. https://doi.org/10.3390/bios12020107
APA StyleLee, J. K., Suh, H. N., Yoon, S. H., Lee, K. H., Ahn, S. Y., Kim, H. J., & Kim, S. H. (2022). Non-Destructive Monitoring via Electrochemical NADH Detection in Murine Cells. Biosensors, 12(2), 107. https://doi.org/10.3390/bios12020107