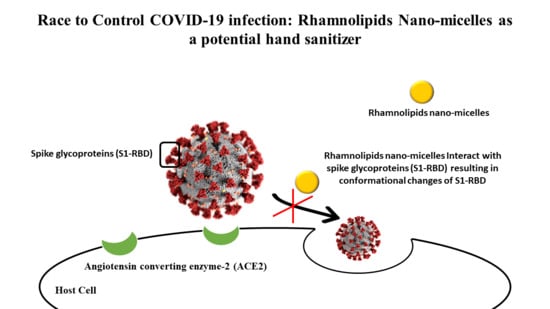
Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer
Abstract
:1. Introduction
2. Results
2.1. Isolation and Identification of Bacterial Strain
2.2. Production and Characterization of Rha(s)
2.2.1. Characterization of Rha(s)
2.3. Preparation and Characterization of Rhamnolipids Nano-Micelles
2.4. Antibacterial Activity of Rhamnolipids Nano-Micelles
2.5. Docking Studies
2.5.1. Molecular Docking
2.5.2. Computational Membrane Permeability and Mode of Action of Rhamnolipids
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Isolation of Biosurfactant Producing Bacterial Strain
4.2.2. Identification of Biosurfactant Producing Bacterial Strain
4.2.3. Production and Characterization of Rha(s)
Production of Rha(s)
Characterization of Rha(s)
4.2.4. Preparation and Characterization of Rha(s) Nano-Micelles
4.2.5. The Antibacterial Activity of Rha(s)
4.2.6. Docking Study
In Silico Molecular Modelling
4.2.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Kratzel, A.; Todt, D.; V’kovski, P.; Steiner, S.; Gultom, M.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Inactivation of Severe Acute Respiratory Syndrome Coronavirus 2 by WHO-Recommended Hand Rub Formulations and Alcohols. Emerg. Infect. Dis. 2020, 26, 1592–1595. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Otter, J.A.; Donskey, C.; Yezli, S.; Douthwaite, S.; Goldenberg, S.D.; Weber, D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowell, S.F.; Simmerman, J.M.; Erdman, D.D.; Wu, J.-S.J.; Chaovavanich, A.; Javadi, M.; Yang, J.-Y.; Anderson, L.J.; Tong, S.; Ho, M.S. Severe Acute Respiratory Syndrome Coronavirus on Hospital Surfaces. Clin. Infect. Dis. 2004, 39, 652–657. [Google Scholar] [CrossRef] [Green Version]
- Emami, A.; Javanmardi, F.; Keshavarzi, A.N.P. Hidden threat lurking behind the alcohol sanitizers in CoVID-19 outbreak. Dermatol. Ther. 2020, 33. [Google Scholar] [CrossRef]
- Slaughter, R.J.; Mason, R.W.; Beasley, D.M.G.; Vale, J.A.; Schep, L.J. Isopropanol poisoning. Clin. Toxicol. 2014, 52, 470–478. [Google Scholar] [CrossRef]
- Mahmood, A.; Eqan, M.; Pervez, S.; Ahmed, H.; Bari, A. COVID-19 and frequent use of hand sanitizers; human health and environmental hazards by exposure pathways. Sci. Total Environ. 2020, 742, 1–7. [Google Scholar] [CrossRef]
- Vogel, L. Hand sanitizers may increase norovirus risk. CMAJ 2011, 183, 799–800. [Google Scholar] [CrossRef] [Green Version]
- Blaney, D.D.; Daly, E.R.; Kirkland, K.B.; Tongren, J.E.; Kelso, P.T.; Talbot, E.A. Use of alcohol-based hand sanitizers as a risk factor for norovirus outbreaks in long-term care facilities in northern New England: December 2006 to March 2007. Am. J. Infect. Control 2011, 39, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shen, C.; Long, X.; Zhang, G.; Meng, Q. Rhamnolipids elicit the same cytotoxic sensitivity between cancer cell and normal cell by reducing surface tension of culture medium. Appl. Microbiol. Biotechnol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Pidot, S.J.; Gao, W.; Buultjens, A.H.; Monk, I.R.; Guerillot, R.; Carter, G.P.; Lee, J.Y.H.; Lam, M.M.C.; Grayson, M.L.; Ballard, S.A.; et al. Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Hayat, A.; Munnawar, F. Antibacterial Effectiveness of Commercially Available Hand Sanitizers. Int. J. Biol. Biotech. 2016, 13, 427–431. [Google Scholar]
- Rosenberg, E.; DeLong, E.F.; Thompson, F.; Lory, S.; Stackebrandt, E. The prokaryotes: Applied bacteriology and biotechnology. Prokaryotes Appl. Bacteriol. Biotechnol. 2013, 9783642313, 1–393. [Google Scholar] [CrossRef]
- Kumar, R.; Das, A.J. Rhamnolipid Biosurfactant: Recent Trends in Production and Application; Springer: Singapore, 2018; ISBN 9789811312892. [Google Scholar]
- Johann, S.; Seiler, T.B.; Tiso, T.; Bluhm, K.; Blank, L.M.; Hollert, H. Mechanism-specific and whole-organism ecotoxicity of mono-rhamnolipids. Sci. Total Environ. 2016. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Pinazo, A.; Jauregui, O.; Espuny, M.J.; Infante, M.R.; Manresa, A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 2003. [Google Scholar] [CrossRef] [Green Version]
- Banat, I.M.; Makkar, R.S.; Cameotra, S.S. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 2000, 53, 495–508. [Google Scholar] [CrossRef]
- Marchant, R.; Banat, I.M. Biosurfactants: A sustainable replacement for chemical surfactants? Biotechnol. Lett. 2012, 34, 1597–1605. [Google Scholar] [CrossRef]
- Bharali, P.; Das, S.; Ray, A.; Pradeep Singh, S.; Bora, U.; Kumar Konwar, B.; Singh, C.B.; Sahoo, D. Biocompatibility natural effect of rhamnolipids in bioremediation process on different biological systems at the site of contamination. Bioremediat. J. 2018, 22, 91–102. [Google Scholar] [CrossRef]
- Müller, M.M.; Hörmann, B.; Syldatk, C.; Hausmann, R. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl. Microbiol. Biotechnol. 2010. [Google Scholar] [CrossRef]
- Lotfabad, T.B.; Shahcheraghi, F.; Shooraj, F. Assessment of antibacterial capability of rhamnolipids produced by two indigenous Pseudomonas aeruginosa strains. Jundishapur J. Microbiol. 2013, 6, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Carrazco-Palafox, J.; Rivera-Chavira, B.E.; Adame-Gallegos, J.R.; Rodríguez-Valdez, L.M.; Orrantia-Borunda, E.; Nevárez-Moorillón, G.V. Rhamnolipids from Pseudomonas aeruginosa Rn19a Modifies the Biofilm Formation over a Borosilicate Surface by Clinical Isolates. Coatings 2021, 11, 136. [Google Scholar] [CrossRef]
- Sana, S.; Datta, S.; Biswas, D.; Sengupta, D. Assessment of synergistic antibacterial activity of combined biosurfactants revealed by bacterial cell envelop damage. Biochim. Biophys. Acta Biomembr. 2018, 1860, 579–585. [Google Scholar] [CrossRef]
- Remichkova, M.; Galabova, D.; Roeva, I.; Karpenko, E.; Shulga, A.; Galabov, A.S. Anti-herpesvirus activities of Pseudomonas sp. S-17 rhamnolipid and its complex with alginate. Zeitschrift Fur Naturforsch. Sect. C J. Biosci. 2008, 63, 75–81. [Google Scholar] [CrossRef]
- Abo-Zeid, Y.; Ismail, N.S.; McLean, G.R.; Hamdy, N.M. A Molecular Docking Study Repurposes FDA Approved Iron Oxide Nanoparticles to Treat and Control COVID-19 Infection. Eur. J. Pharm. Sci. 2020, 153, 105465. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.; Malik, Y.S.; Tomar, S. Identification of SARS-CoV-2 Cell Entry Inhibitors by Drug Repurposing Using in silico Structure-Based Virtual Screening Approach. Front. Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Déziel, E.; Lépine, F.; Dennie, D.; Boismenu, D.; Mamer, O.A.; Villemur, R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1440, 244–252. [Google Scholar] [CrossRef]
- Zhao, F.; Shi, R.; Ma, F.; Han, S.; Zhang, Y. Oxygen effects on rhamnolipids production by Pseudomonas aeruginosa. Microb. Cell Fact. 2018, 17, 1–11. [Google Scholar] [CrossRef]
- Zhao, F.; Jiang, H.; Sun, H.; Liu, C.; Han, S.; Zhang, Y. Production of rhamnolipids with different proportions of mono-rhamnolipids using crude glycerol and a comparison of their application potential for oil recovery from oily sludge. RSC Adv. 2019, 9, 2885–2891. [Google Scholar] [CrossRef] [Green Version]
- Pantazaki, A.A.; Papaneophytou, C.P.; Lambropoulou, D.A. Simultaneous polyhydroxyalkanoates and rhamnolipids production by thermus thermophilus HB8. AMB Express 2011, 1, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Leung, S.S.F.; Sindhikara, D.; Jacobson, M.P. Simple Predictive Models of Passive Membrane Permeability Incorporating Size-Dependent Membrane-Water Partition. J. Chem. Inf. Model. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ndlovu, T.; Rautenbach, M.; Vosloo, J.A.; Khan, S.; Khan, W. Characterisation and antimicrobial activity of biosurfactant extracts produced by Bacillus amyloliquefaciens and Pseudomonas aeruginosa isolated from a wastewater treatment plant. AMB Express 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvis, F.G.; Johnson, M.J. A Glyco-lipide Produced by Pseudomonas Aeruginosa. J. Am. Chem. Soc. 1949. [Google Scholar] [CrossRef]
- Tiso, T.; Thies, S.; Müller, M.; Tsvetanova, L.; Carraresi, L.; Bröring, S.; Jaeger, K. Rhamnolipids: Production, Performance, and Application. In Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Production of Fuels and Chemicals; Lee, S.Y., Ed.; Springer: Cham, Switzerland, 2017; ISBN 9783319504360. [Google Scholar]
- Chong, H.; Li, Q. Microbial production of rhamnolipids: Opportunities, challenges and strategies. Microb. Cell Fact. 2017, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [Green Version]
- Nie, M.; Yin, X.; Ren, C.; Wang, Y.; Xu, F.; Shen, Q. Novel rhamnolipid biosurfactants produced by a polycyclic aromatic hydrocarbon-degrading bacterium Pseudomonas aeruginosa strain NY3. Biotechnol. Adv. 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaskatepe, B.; Yildiz, S. Rhamnolipid biosurfactants produced by pseudomonas Species. Brazilian Arch. Biol. Technol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Arutchelvi, J.; Doble, M. Characterization of glycolipid biosurfactant from Pseudomonas aeruginosa CPCL isolated from petroleum-contaminated soil. Lett. Appl. Microbiol. 2010, 51, 75–82. [Google Scholar] [CrossRef]
- Tan, Y.N.; Li, Q. Microbial production of rhamnolipids using sugars as carbon sources. Microb. Cell Fact. 2018, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 2014, 5, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdemir, S.; Yetilmezsoy, K. A mini literature review on sustainable management of poultry abattoir wastes. J. Mater. Cycles Waste Manag. 2020, 22, 11–21. [Google Scholar] [CrossRef]
- IA, B. Traditional Methods of Carcass Disposal: A Review. J. Dairy Vet. Anim. Res. 2017, 5, 21–27. [Google Scholar] [CrossRef]
- Makkar, R.S.; Cameotra, S.S.; Banat, I.M. Advances in utilization of renewable substrates for biosurfactant production. AMB Express 2011, 1, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Sabturani, N.; Latif, J.; Radiman, S.; Hamzah, A. Analisis spektroskopik ramnolipid yang dihasilkan oleh P. aeruginosa UKMP14T. Malays. J. Anal. Sci. 2016, 20, 31–43. [Google Scholar] [CrossRef]
- Rikalović, M.G.; Gojgić-Cvijović, G.; Vrvić, M.M.; Karadžić, I. Production and characterization of rhamnolipids from Pseudomonas aeruginosa san-ai. J. Serbian Chem. Soc. 2012, 77, 27–42. [Google Scholar] [CrossRef]
- Abalos, A.; Pinazo, A.; Infante, M.R.; Casals, M.; García, F.; Manresa, A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir 2001. [Google Scholar] [CrossRef]
- Nitschke, M.; Paulista, U.E.; Nitschke, M.; Costa, S.G.V.A.O.; Haddad, R.; Gonc, L.A.G.; Eberlin, M.N.; Contiero, J. Oil Wastes as Unconventional Substrates for Rhamnolipid Biosurfactant Production by Pseudomonas aeruginosa LBI Oil Wastes as Unconventional Substrates for Rhamnolipid Biosurfactant Production by Pseudomonas aeruginosa LBI. Biotechnol. Prog. 2005, 3, 1562–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, K.; Li, H.; Abo-Zeid, Y.; Fatimah; Williams, G.R. The effect of molecular properties on active ingredient release from electrospun eudragit fibers. Pharmaceutics 2018, 10, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, Z.; Abo-zeid, Y.; Bear, J.C.; Davies, G.; Lei, X.; Williams, G.R. SiO2-coated layered gadolinium hydroxides for simultaneous drug delivery and magnetic resonance imaging. J. Solid State Chem. 2020. [Google Scholar] [CrossRef]
- Abo-zeid, Y.; Garnett, M.C. Polymer nanoparticle as a delivery system for ribavirin: Do nanoparticle avoid uptake by Red Blood Cells? J. Drug Deliv. Sci. Technol. 2020, 56, 101552. [Google Scholar] [CrossRef]
- Chintagunta, A.D.; Krishna, M.S.; Nalluru, S.; Sampath, S.K. Nanotechnology: An emerging approach to combat COVID-19. Emergent Mater. 2021. [Google Scholar] [CrossRef]
- Rangayasami, A.; Kannan, K.; Murugesan, S.; Radhika, D.; Sadasivuni, K.K.; Reddy, K.R.; Raghu, A.V. Influence of nanotechnology to combat against COVID-19 for global health emergency: A review. Sens. Int. 2021, 2, 100079. [Google Scholar] [CrossRef]
- Campos, E.V.R.; Pereira, A.E.S.; De Oliveira, J.L.; Carvalho, L.B.; Guilger-Casagrande, M.; De Lima, R.; Fraceto, L.F. How can nanotechnology help to combat COVID-19? Opportunities and urgent need. J. Nanobiotechnol. 2020, 18, 1–23. [Google Scholar] [CrossRef]
- Abo-zeid, Y.; Urbanowicz, R.A.; Thomsonb, B.J.; William, L.; Irvingb, A.W.T.; Garnett, M.C. Enhanced nanoparticle uptake into virus infected cells: Could nanoparticles be useful in antiviral therapy? Int. J. Pharm. 2018, 547, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Abo-zeid, Y.; Williams, G.R.; Touabi, L.; Mclean, G.R. An investigation of rhinovirus infection on cellular uptake of poly (glycerol-adipate) nanoparticles. Int. J. Pharm. 2020, 119826. [Google Scholar] [CrossRef] [PubMed]
- Abo-zeid, Y.; Mantovani, G.; Irving, W.L.; Garnett, M.C. Synthesis of nucleoside-boronic esters hydrophobic pro-drugs: A possible route to improve hydrophilic nucleoside drug loading into polymer nanoparticles. J. Drug Deliv. Sci. Technol. 2018, 46, 354–364. [Google Scholar] [CrossRef]
- AlMatar, M.; Makky, E.A.; Var, I.; Koksal, F. The Role of Nanoparticles in the Inhibition of Multidrug-resistant Bacteria and Biofilms. Curr. Drug Deliv. 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-strategies to fight multidrug resistant bacteria-”A Battle of the Titans”. Front. Microbiol. 2018, 9, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ssekatawa, K.; Byarugaba, D.K.; Kato, C.D.; Ejobi, F.; Tweyongyere, R.; Lubwama, M.; Kirabira, J.B.; Wampande, E.M. Nanotechnological solutions for controlling transmission and emergence of antimicrobial-resistant bacteria, future prospects, and challenges: A systematic review. J. Nanoparticle Res. 2020, 22. [Google Scholar] [CrossRef]
- Pepić, I.; Lovrić, J.; Hafner, A.; Filipović-Grčić, J. Powder form and stability of Pluronic mixed micelle dispersions for drug delivery applications. Drug Dev. Ind. Pharm. 2014, 40, 944–951. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Putri, D.C.A.; Dwiastuti, R.; Marchaban, M.; Nugroho, A.K. Optimasi Suhu Pencampuran Dan Durasi Sonikasi Dalam Pembuatan Liposom. J. Pharm. Sci. Community 2017, 14, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Piazzini, V.; D’Ambrosio, M.; Luceri, C.; Cinci, L.; Landucci, E.; Bilia, A.R.; Bergonzi, M.C. Formulation of nanomicelles to improve the solubility and the oral absorption of silymarin. Molecules 2019, 24, 1688. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Yang, X.P.; Ma, L.Z. Analysis of biosurfactants from industrially viable Pseudomonas strain isolated from crude oil suggests how rhamnolipids congeners affect emulsification property and antimicrobial activity. Front. Microbiol. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bharali, P.; Saikia, J.P.; Ray, A.; Konwar, B.K. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: A novel chemotaxis and antibacterial agent. Colloids Surfaces B Biointerfaces 2013, 103, 502–509. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Ferreira, J.; Vieira, E.A.; Nitschke, M. The antibacterial activity of rhamnolipid biosurfactant is pH dependent. Food Res. Int. 2019, 116, 737–744. [Google Scholar] [CrossRef]
- Díaz De Rienzo, M.A.; Kamalanathan, I.D.; Martin, P.J. Comparative study of the production of rhamnolipid biosurfactants by B. thailandensis E264 and P. aeruginosa ATCC 9027 using foam fractionation. Process Biochem. 2016, 51, 820–827. [Google Scholar] [CrossRef]
- Otzen, D.E. Biosurfactants and surfactants interacting with membranes and proteins: Same but different? Biochim. Biophys. Acta Biomembr. 2017, 1859, 639–649. [Google Scholar] [CrossRef]
- Kaczorek, E.; Chrzanowski, Ł.; Pijanowska, A.; Olszanowski, A. Yeast and bacteria cell hydrophobicity and hydrocarbon biodegradation in the presence of natural surfactants: Rhamnolipides and saponins. Bioresour. Technol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.M.; Shi, J.G.; Yuan, X.Z.; Liu, J.; Zhang, Z.B.; Huang, G.H.; Li, J.B.; Xi, B.D.; Liu, H.L. Effects of Tween 80 and rhamnolipid on the extracellular enzymes of Penicillium simplicissimum isolated from compost. Enzyme Microb. Technol. 2006. [Google Scholar] [CrossRef]
- Zeng, G.; Liu, Z.; Zhong, H.; Li, J.; Yuan, X.; Fu, H.; Ding, Y.; Wang, J.; Zhou, M. Effect of monorhamnolipid on the degradation of n-hexadecane by Candida tropicalis and the association with cell surface properties. Appl. Microbiol. Biotechnol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Liu, Z.; Zhong, H.; Zeng, G.; Liu, G.; Yu, M.; Liu, Y.; Yang, X.; Li, Z.; Fang, Z.; et al. Effects of rhamnolipids on microorganism characteristics and applications in composting: A review. Microbiol. Res. 2017, 200, 33–44. [Google Scholar] [CrossRef]
- Chaudhary, N.K.; Chaudhary, N.; Dahal, M.; Guragain, B.; Rai, S.; Chaudhary, R.; Sachin, K.M.; Lamichhane-Khadka, R.; Bhattarai, A. Fighting the SARS CoV-2 (COVID-19) Pandemic with Soap. Preprints 2020, 060, 1–19. [Google Scholar]
- Smith, M.L.; Gandolfi, S.; Coshall, P.M.; Rahman, P.K.S.M. Biosurfactants: A Covid-19 Perspective. Front. Microbiol. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Roth, Y.; Opatowski, E.; Lichtenberg, D.; Kozlov, M.M. Phase behavior of dilute aqueous solutions of lipid-surfactant mixtures: Effects of finite size of micelles. Langmuir 2000, 16, 2052–2061. [Google Scholar] [CrossRef]
- Kragh-hansen, U.; Maire, M.; Møller, J.V. The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophys. J. 1998, 75, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Koynova, R.; Tenchov, B. Interactions of surfactants and fatty acids with lipids. Curr. Opin. Colloid Interface Sci. 2001, 6, 277–286. [Google Scholar] [CrossRef]
- Fredriksson, N.J.; Hermansson, M.; Wilén, B.-M. The Choice of PCR Primers Has Great Impact on Assessments of Bacterial Community Diversity and Dynamics in a Wastewater Treatment Plant. PLoS ONE 2013, 8, e76431. [Google Scholar] [CrossRef]
- Rahman, P.K.S.M.; Pasirayi, G.; Auger, V.; Ali, Z. Production of rhamnolipid biosurfactants by Pseudomonas aeruginosa DS10-129 in a microfluidic bioreactor. Biotechnol. Appl. Biochem. 2010, 55, 45–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benincasa, M.; Abalos, A.; Oliveira, I.; Manresa, A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2004, 85, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing Supplement M100S; CLSI: Wayne, PA, USA, 2016; ISBN 1-56238-923-8. [Google Scholar]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Rha(s) Congeners | m/z | |||||||
|---|---|---|---|---|---|---|---|---|
| Mol f | Mol wt | [M-H]− | [M+H]+ | [M+Na]+ | [M+K]+ | [M-H+Na2]+ | % Abundance | |
| Mono-Rhamnolipid (Rha(s)1) Congeners | ||||||||
| R-C8 | C14H26O7 | 306 | 351 | 12.8 | ||||
| R-C8:1 | C14H24O7 | 304 | 327 | 25.2 | ||||
| R-C8:2 | C14H22O7 | 302 | 325 | 29.4 | ||||
| R-C9:1 | C15H26O7 | 318 | 341 | 0.3 | ||||
| R-C10 | C16H30O7 | 334 | 357 | 379 | 13.6 | |||
| R-C10:2 | C16H26O7 | 330 | 353 | 0.12 | ||||
| R-C12 | C18H34O7 | 362 | 385 | 0.24 | ||||
| R-C12:2 | C18H30O7 | 358 | 359 | 381 | 2.28 | |||
| R-C13 | C19H36O7 | 376 | 421 | 0.56 | ||||
| R-C13:2 | C19H32O7 | 372 | 395 | 0.8 | ||||
| R-C14 | C20H38O7 | 390 | 413 | 0.06 | ||||
| R-C15 | C21H40O7 | 404 | 443 | 0.24 | ||||
| R-C8 -C12, R-C9-C11, R-C10-C10, R-C12 -C8, R-C11-C9 | C26H48O9 | 504 | 503 | 527 | 543 | 6.8 | ||
| R-C8-C14, R-C9-C13, R-C10-C12, R-C11-C11 | C28H52O9 | 532 | 531 | 577 | 0.52 | |||
| R-C8-C14:1, R-C9-C13:1, R-C10-C12:1, R-C11-C11:1, R-C8:1-C14, R-C9:1-C13, R-C10:1-C12, R-C11:1-C11 | C28H50O9 | 530 | 553 | 0.6 | ||||
| R-C11-C16, R-C12-C15, R-C13-C14 | C33H62O9 | 602 | 603 | 641 | 0.52 | |||
| R-C14-C16:2, R-C15-C15:2, R-C14:2-C16, R-C15:2-C15 | C36H64O9 | 640 | 641 | 0.8 | ||||
| Di-rhamnolipid (Rha(s)2) congeners | ||||||||
| R-R-C12:1 | C24H42O11 | 506 | 551 | 0.22 | ||||
| R-R-C16:1 | C28H50O11 | 562 | 601 | 0.62 | ||||
| R-R-C8-C10:2, R-R-C9-C9:2, R-R-C8:2-C10, R-R-C9:2-C9 | C30H50O13 | 618 | 641 | 657 | 1.42 | |||
| R-R-C8 -C12, R-R-C9 -C11, R-R-C10 -C10, R-R-C12-C8, R-R-C11 -C9, | C32H58O13 | 650 | 649 | 673 | 1.6 | |||
| R-R-C16-C16:2, R-R-C16:2-C16 | C44H78O13 | 815 | 816 | 0.03 | ||||
| Mol F, Molecular formula Mol wt, Molecular weight R, Rhamnose | ||||||||
| Concentrations of Rhamnolipids (mg mL−1) | Particle Size (D nm ± SD) | Polydispersity Index (PDI) | Zeta Potential (mv ± SD) |
|---|---|---|---|
| 1 | 274 ± 50 | 0.55 | −50.4 ± 1.7 |
| 5 | 164 ± 1 | 0.30 | −62.07 ± 3.8 |
| 10 | 169 ± 10 | 0.27 | −66.77 ± 2.62 |
| Bacterial Strain | Concentration of Rha(s) (mg mL−1) | |||
|---|---|---|---|---|
| MIC | 1 | 5 | 10 | |
| Corresponding Zone of Inhibition (mm) ± SD | ||||
| Gram-positive bacteria | ||||
| Streptococcus pneumoniae | 0.031 | 9.6 ± 1.2 | 16.5 ± 1 | 23 ± 2 |
| Staphylococcus aureus | 0.031 | 17.8 ± 0.76 | 25 ± 1 | 30 ± 1.5 |
| Gram-negative bacteria | ||||
| Salmonella Montevideo | >0.5 | 7.1 ± 1 | 15 ± 1 | 21 ± 1.5 |
| Salmonella Typhimurium | >0.5 | 6.8 ± 0.76 | 12.1 ± 1.2 | 18.1 ± 1.7 |
| Ligands | Binding Free Energy (kcal/mol) | Total Intermolecular Energy (kcal/mol) | Interacting Amino Acids | Hydrogen Bonds |
|---|---|---|---|---|
| Spike glycoproteins | ||||
| Rha(s)1 | −45 | 14.7 | Gln 52 and Thr 739 | 3H bonds |
| Rha(s)2 | −44.6 | 11.8 | Gly 757 | 1H bonds |
| EndoRNAse | ||||
| Rha(s)1 | −61 | 20.5 | Glu 41, Glu 44 and Glu 266 | 5H bonds |
| Rha(s)2 | −53.9 | 11.5 | Asp, Glu44, and Lys 46 | 4H bonds |
| Helicase | ||||
| Rha(s)1 | −66.4 | 14.2 | Gln 537, Glu 375, and Lys 288 | 3H bonds |
| Rha(s)2 | −35.5 | 5.7 | Asp542, Glu 540, and Lys 508 | 4H bonds |
| RNA-dependent RNA polymerase | ||||
| Rha(s)1 | −62.1 | 17.3 | Arg 555, Arg 624, Asp 618, Thr 556 and Lys 621 | 7H bonds |
| Rha(s)2 | −55.8 | 13.7 | Arg 555, Arg 624, Asp 618, Thr 556, Arg 553, and Lys 621 | 6H bonds |
| Protease | ||||
| Rha(s)1 | −77 | 22.1 | Glu 288, Glu 290, Leu 282 and Lys 5 | 7H bonds |
| Rha(s)2 | −61.1 | 12.1 | Glu 288, Glu 290, Gly 283 and Lys 5 | 6H bonds |
| Ligand | Membrane Permeability Prediction | |||||
|---|---|---|---|---|---|---|
| 1 Membrane *dG Insert | 2 Membrane HDLD | 3 Membrane GB | 4 Membrane State Penalty | 5 Log Perm RRCK (cm/s) | Membrane Energy | |
| Rha(s)1 | 9.909 | 5.516 | −3.033 | 9.909 | −5.854 | 13.416 |
| Rha(s)2 | 6.004 | 1.610 | −6.789 | 6.004 | −5.466 | −1.146 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakkar, M.R.; Faraag, A.H.I.; Soliman, E.R.S.; Fouda, M.S.; Sarguos, A.M.M.; McLean, G.R.; Hebishy, A.M.S.; Elkhouly, G.E.; Raya, N.R.; Abo-zeid, Y. Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer. Antibiotics 2021, 10, 751. https://doi.org/10.3390/antibiotics10070751
Bakkar MR, Faraag AHI, Soliman ERS, Fouda MS, Sarguos AMM, McLean GR, Hebishy AMS, Elkhouly GE, Raya NR, Abo-zeid Y. Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer. Antibiotics. 2021; 10(7):751. https://doi.org/10.3390/antibiotics10070751
Chicago/Turabian StyleBakkar, Marwa Reda, Ahmed Hassan Ibrahim Faraag, Elham R. S. Soliman, Manar S. Fouda, Amir Mahfouz Mokhtar Sarguos, Gary R. McLean, Ali M. S. Hebishy, Gehad E. Elkhouly, Nermeen R. Raya, and Yasmin Abo-zeid. 2021. "Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer" Antibiotics 10, no. 7: 751. https://doi.org/10.3390/antibiotics10070751
APA StyleBakkar, M. R., Faraag, A. H. I., Soliman, E. R. S., Fouda, M. S., Sarguos, A. M. M., McLean, G. R., Hebishy, A. M. S., Elkhouly, G. E., Raya, N. R., & Abo-zeid, Y. (2021). Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer. Antibiotics, 10(7), 751. https://doi.org/10.3390/antibiotics10070751