Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Prediction of Toxicity
2.3. Biological Evaluation
2.3.1. Antibacterial Activity
2.3.2. Antifungal Activity
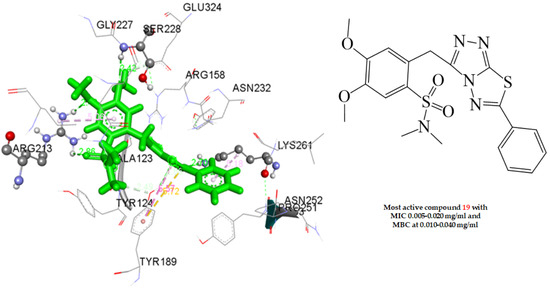
2.4. Docking Studies
2.4.1. Docking to Antibacterial Targets
2.4.2. Docking to Antifungal Targets
2.5. Search for Structural Analogs
2.6. In-Silico Predictive Studies
2.7. Cytotoxicity Assays
3. Materials and Methods
3.1. Biological Valuation
3.1.1. Antimicrobial Activity
3.1.2. Inhibition of Biofilm Formation
3.2. Antifungal Activity
3.3. Statistical Analysis
3.4. Docking
3.5. Chemical Similarity Assessment
3.6. In-Silico Predictive Studies
3.7. Cytotoxicity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, R.; Liu, J.; Li, M.; Huang, J.; Sun, J.; Qu, H. Epidemiology and antimicrobial resistance among commonly encountered bacteria associated with infections and colonization in intensive care units in a university-affiliated hospital in Shanghai. J. Microbiol. Immunol. Infect. 2014, 47, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Pfeltz, R.; Wilkinson, B. The Escalating Challenge of Vancomycin Resistance in Staphylococcus aureus. Curr. Drug Targets Infect. Disord. 2004, 4, 273–294. [Google Scholar] [CrossRef]
- Roberts, M.C. Distribution of Macrolide, Lincosamide, Streptogramin, Ketolide and Oxazolidinone (MLSKO) Resistance Genes in Gram-negative Bacteria. Curr. Drug Targets Infect. Disord. 2004, 4, 207–215. [Google Scholar] [CrossRef]
- Dessen, A.; Di Guilmi, A.M.; Vernet, T.; Dideberg, O. Molecular mechanisms of antibiotic resistance in gram-positive pathogens. Curr. Drug Targets Infect. Disord. 2001, 1, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P. Surveillance of antibiotic resistance. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140080. [Google Scholar] [CrossRef] [Green Version]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of Kibdelomycin, A Potent New Class of Bacterial Type II Topoisomerase Inhibitor by Chemical-Genetic Profiling in Staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef] [Green Version]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef]
- Maillard, L.T.; Bertout, S.; Quinonéro, O.; Akalin, G.; Turan-Zitouni, G.; Fulcrand, P.; Demirci, F.; Martinez, J.; Masurier, N. Synthesis and anti-Candida activity of novel 2-hydrazino-1,3-thiazole derivatives. Bioorg. Med. Chem. Lett. 2013, 23, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Farowski, F.; Vehreschild, J.J.; A Cornely, O. Posaconazole: A next-generation triazole antifungal. Future Microbiol. 2007, 2, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Keri, R.S.; Patil, S.A.; Budagumpi, S.; Nagaraja, B.M. Triazole: A Promising Antitubercular Agent. Chem. Biol. Drug Des. 2015, 86, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Altimari, J.M.; Hockey, S.C.; Boshoff, H.I.; Sajid, A.; Henderson, L.C. Novel 1,4-Substituted-1,2,3-Triazoles as Antitubercular Agents. ChemMedChem 2015, 10, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, M.H.; Subhedar, D.D.; Nawale, L.; Sarkar, D.; Khan, F.A.K.; Sangshetti, J.N.; Shingate, B.B. 1,2,3-Triazole derivatives as antitubercular agents: Synthesis, biological evaluation and molecular docking study. MedChemComm 2015, 6, 1104–1116. [Google Scholar] [CrossRef]
- Ahmadi, F.; Ghayahbashi, M.; Sharifzadeh, M.; Alipoiur, E.; Ostad, S.; Vosooghi, M.; Khademi, H.; Amini, M. Synthesis and Evaluation of Anti-inflammatory and Analgesic Activities of New 1,2,4-triazole Derivatives. Med. Chem. 2014, 11, 69–76. [Google Scholar] [CrossRef]
- Khanage, S.G.; Raju, A.; Mohite, P.B.; Pandhare, R.B. Analgesic Activity of Some 1,2,4-Triazole Heterocycles Clubbed with Pyrazole, Tetrazole, Isoxazole and Pyrimidine. Adv. Pharm. Bull. 2013, 3, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Grewal, A.S.; Lather, V.; Pandita, D.; Dalal, R. Synthesis, Docking and Anti-inflammatory Activity of Triazole Amine Derivatives as Potential Phosphodiesterase-4 Inhibitors. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2017, 16, 58–67. [Google Scholar] [CrossRef]
- Liu, C.; Bian, M.; Yu, L.; Wei, C. Synthesis and Anti-Inflammatory Activity Evaluation of 5-(1-Benzyl-1H-[1,2,3]Triazol-4-yl)-4-Phenyl- 4H-[1,2,4]Triazole-3-Thiol Derivatives. Indian J. Pharm. Educ. Res. 2018, 52, 505–513. [Google Scholar] [CrossRef] [Green Version]
- El-Sherief, H.A.; Youssif, B.G.; Bukhari, S.N.A.; Abdel-Aziz, M.; Abdel-Rahman, H. Novel 1,2,4-triazole derivatives as potential anticancer agents: Design, synthesis, molecular docking and mechanistic studies. Bioorg. Chem. 2018, 76, 314–325. [Google Scholar] [CrossRef]
- Song, M.-X.; Deng, X.-Q. Recent developments on triazole nucleus in anticonvulsant compounds: A review. J. Enzym. Inhib. Med. Chem. 2018, 33, 453–478. [Google Scholar] [CrossRef] [Green Version]
- Sari, S.; Kaynak, F.B.; Dalkara, S. Synthesis and anticonvulsant screening of 1,2,4-triazole derivatives. Pharmacol. Rep. 2018, 70, 1116–1123. [Google Scholar] [CrossRef]
- Yar, M.S.; Sharma, P.C. Recent Advances and Future Perspectives of Triazole Analogs as Promising Antiviral Agents. Mini-Rev. Med. Chem. 2011, 11, 84–96. [Google Scholar] [CrossRef]
- Aouad, M.R.; Mayaba, M.M.; Naqvi, A.; Bardaweel, S.; Al-Blewi, F.F.; Messali, M.; Rezki, N. Design, synthesis, in silico and in vitro antimicrobial screening of novel 1,2,4-triazoles carrying 1,2,3-trizole scaffold with lipophilic side chain tether. Chem. Cent. J. 2017, 11, 117. [Google Scholar] [CrossRef] [Green Version]
- Aouad, M.R.; Messali, M.; Rezki, N.; Ali, A.A.-S.; Lesimple, A. Synthesis and characterization of some novel 1,2,4-triazoles, 1,3,4-thiadiazoles and Schiff bases incorporating imidazole moiety as potential antimicrobial agents. Acta Pharm. 2015, 65, 117–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, J.-R.; Liu, H.-B.; Jeyakkumar, P.; Gopala, L.; Li, S.; Geng, R.-X.; Zhou, C.-H. Design, synthesis and biological evaluation of novel Schiff base-bridged tetrahydroprotoberberine triazoles as a new type of potential antimicrobial agents. MedChemComm 2017, 8, 907–916. [Google Scholar] [CrossRef]
- Bektaș, H.; Karrali, N.; Sahin, D.; Demirbaș, A.; Karaoglu, S.A.; Demirbaș, N. Synthesis and antimicrobial activities of some new 1,2,4-triazole derivative. Molecules 2010, 15, 2427–2438. [Google Scholar] [CrossRef] [Green Version]
- Aburahama, G.; Hassan, H.; Ezelarab, H.; Abbas, S.H.; El-Baky, R.A. Design, Synthesis and Antifungal Activity of 1,2,4-Triazole/or 1,3,4- Oxadiazole-ciprofloxacin hybrids. J. Adv. Biomed. Pharm. Sci. 2018, 1, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Chai, X.; Wang, Y.; Cao, Y.-B.; Zhang, J.; Wu, Q.; Zhang, D.; Jiang, Y.; Yan, T.; Sun, Q.-Y. Triazole derivatives with improved in vitro antifungal activity over azole drugs. Drug Des. Dev. Ther. 2014, 8, 383–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadeghpour, H.; Khabnadideh, S.; Zomorodian, K.; Pakshir, K.; Hoseinpour, K.; Javid, N.; Faghih-Mirzaei, E.; Rezaei, Z. Design, Synthesis, and Biological Activity of New Triazole and Nitro-Triazole Derivatives as Antifungal Agents. Molecules 2017, 22, 1150. [Google Scholar] [CrossRef]
- Dai, Z.-C.; Chen, Y.-F.; Zhang, M.; Li, S.-K.; Yang, T.-T.; Shen, L.; Wang, J.-X.; Qian, S.-S.; Zhu, H.-L.; Ye, Y.-H. Synthesis and antifungal activity of 1,2,3-triazole phenylhydrazone derivatives. Org. Biomol. Chem. 2014, 13, 477–486. [Google Scholar] [CrossRef]
- Noolvi, M.N.; Patel, H.M.; Kamboj, S.; Cameotra, S.S. Synthesis and antimicrobial evaluation of novel 1,3,4-thiadiazole derivatives of 2-(4-formyl-2-methoxyphenoxy) acetic acid. Arab. J. Chem. 2016, 9, S1283–S1289. [Google Scholar] [CrossRef] [Green Version]
- Tahtaci, H.; Karacık, H.; Ece, A.; Er, M.; Şeker, M.G. Design, Synthesis, SAR and Molecular Modeling Studies of Novel Imidazo[2,1-b][1,3,4]Thiadiazole Derivatives as Highly Potent Antimicrobial Agents. Mol. Inform. 2018, 37, 10. [Google Scholar] [CrossRef] [PubMed]
- Serban, G.; Stanasel, O.; Serban, E.; Bota, S. 2-Amino-1,3,4-thiadiazole as a potential scaffold for promising antimicrobial agents. Drug Des. Dev. Ther. 2018, 12, 1545–1566. [Google Scholar] [CrossRef] [Green Version]
- Er, M.; Ergüven, B.; Tahtaci, H.; Onaran, A.; Karakurt, T.; Ece, A. Synthesis, characterization, preliminary SAR and molecular docking study of some novel substituted imidazo[2,1-b][1,3,4]thiadiazole derivatives as antifungal agents. Med. Chem. Res. 2017, 26, 615–630. [Google Scholar] [CrossRef]
- Yan, S.-L.; Yang, M.-Y.; Sun, Z.-H.; Min, L.-J.; Tan, C.-X.; Weng, J.-Q.; Wu, H.-K.; Liu, X.-H. Synthesis and Antifungal Activity of 1,2,3-thiadiazole Derivatives Containing 1,3,4-thiadiazole Moiety. Lett. Drug Des. Discov. 2014, 11, 940–943. [Google Scholar] [CrossRef]
- Fesatidou, M.; Zagaliotis, P.; Camoutsis, C.; Petrou, A.; Eleftheriou, P.; Tratrat, C.; Haroun, M.; Geronikaki, A.; Ciric, A.; Sokovic, M. 5-Adamantan thiadiazole-based thiazolidinones as antimicrobial agents. Design, synthesis, molecular docking and evaluation. Bioorg. Med. Chem. 2018, 26, 4664–4676. [Google Scholar] [CrossRef]
- Cui, Z.-N.; Li, Y.-S.; Hu, D.-K.; Tian, H.; Jiang, J.-Z.; Wang, Y.; Yan, X.-J. Synthesis and fungicidal activity of novel 2,5-disubstituted-1,3,4- thiadiazole derivatives containing 5-phenyl-2-furan. Sci. Rep. 2016, 6, 20204. [Google Scholar] [CrossRef] [Green Version]
- Levent, S.; Çavuşoğlu, B.K.; Sağlık, B.N.; Osmaniye, D.; Çevik, U.A.; Atlı, Ö.; Özkay, Y.; Kaplancıklı, Z.A. Synthesis of Oxadiazole-Thiadiazole Hybrids and Their Anticandidal Activity. Molecules 2017, 22, 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.K.; Sharma, S.; Vaidya, A.; Ravichandran, V.; Agrawal, R.K. 1,3,4-Thiadiazole and its Derivatives: A Review on Recent Progress in Biological Activities. Chem. Biol. Drug Des. 2013, 81, 557–576. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Taya, P. A review on the various biological activities of thiadiazole. Int. J. Pharm. Sci. 2015, 7, 39–47. [Google Scholar]
- Can, N.Ö.; Can, Ö.D.; Osmaniye, D.; Özkay, Ü.D. Synthesis of Some Novel Thiadiazole Derivative Compounds and Screening Their Antidepressant-Like Activities. Molecules 2018, 23, 716. [Google Scholar] [CrossRef] [Green Version]
- Hussein, E.M.; Al-Rooqi, M.M.; El-Galil, S.M.A.; Ahmed, S.A. Design, synthesis, and biological evaluation of novel N4-substituted sulfonamides: Acetamides derivatives as dihydrofolate reductase (DHFR) inhibitors. BMC Chem. 2019, 13, 91. [Google Scholar] [CrossRef]
- Kwon, Y.; Song, J.; Lee, H.; Kim, E.-Y.; Lee, K.; Lee, S.K.; Kim, S. Design, Synthesis, and Biological Activity of Sulfonamide Analogues of Antofine and Cryptopleurine as Potent and Orally Active Antitumor Agents. J. Med. Chem. 2015, 58, 7749–7762. [Google Scholar] [CrossRef] [Green Version]
- Okolotowicz, K.J.; Dwyer, M.; Ryan, D.; Cheng, J.; Cashman, E.A.; Moore, S.; Mercola, M.; Cashman, J.R. Novel tertiary sulfonamides as potent anti-cancer agents. Bioorg. Med. Chem. 2018, 26, 4441–4451. [Google Scholar] [CrossRef] [PubMed]
- Gokcen, T.; Gulcin, I.; Ozturk, T.; Goren, A.C. A class of sulfonamides as carbonic anhydrase I and II inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 180–188. [Google Scholar] [CrossRef]
- Ferraroni, M.; Cornelio, B.; Sapi, J.; Supuran, C.T.; Scozzafava, A. Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms. Inorg. Chim. Acta 2018, 470, 128–132. [Google Scholar] [CrossRef]
- Bua, S.; Mannelli, L.D.C.; Vullo, D.; Ghelardini, C.; Bartolucci, G.; Scozzafava, A.; Supuran, C.T.; Carta, F. Design and Synthesis of Novel Nonsteroidal Anti-Inflammatory Drugs and Carbonic Anhydrase Inhibitors Hybrids (NSAIDs–CAIs) for the Treatment of Rheumatoid Arthritis. J. Med. Chem. 2017, 60, 1159–1170. [Google Scholar] [CrossRef]
- Eze, F.U.; Okoro, U.C.; Ugwu, D.; Okafor, S.N. Biological Activity Evaluation of Some New Benzenesulphonamide Derivatives. Front. Chem. 2019, 7, 634. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Vozzolo, L.; Mok, B.J.; Lee, C.C.; Fitzmaurice, R.J.; Caddick, S.; Fassati, A. Inhibition of HIV-1 Replication by Isoxazolidine and Isoxazole Sulfonamides. Chem. Biol. Drug Des. 2010, 75, 461–474. [Google Scholar] [CrossRef]
- Che, Z.; Tian, Y.; Liu, S.; Hu, M.; Chen, G. Synthesis and in vitro anti-HIV-1 evaluation of some N-arylsulfonyl-3-formylindoles. Braz. J. Pharm. Sci. 2018, 54. [Google Scholar] [CrossRef] [Green Version]
- Ghorab, M.M.; Soliman, A.M.; Alsaid, M.S.; Askar, A. Synthesis, antimicrobial activity and docking study of some novel 4-(4,4-dimethyl-2,6-dioxocyclohexylidene)methylamino derivatives carrying biologically active sulfonamide moiety. Arab. J. Chem. 2020, 13, 545–556. [Google Scholar] [CrossRef]
- Tačić, A.; Nikolić, V.; Nikolić, L.; Savić, I. Antimicrobial sulfonamide drugs. Adv. Technol. 2017, 6, 58–71. [Google Scholar] [CrossRef] [Green Version]
- Genç, Y.; Özkanca, R.; Bekdemir, Y. Antimicrobial activity of some sulfonamide derivatives on clinical isolates of Staphylococus aureus. Ann. Clin. Microbiol. Antimicrob. 2008, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Beheshtimaal, K.; Khazaeili, T.; Asakere, N.; Mousavi, F.; Massah, A.R.; Assakere, N. Synthesis of Some Novel Sulfonamide-imines as Potential Antimicrobial Agents. Lett. Org. Chem. 2018, 15, 2. [Google Scholar] [CrossRef]
- Qadir, M.A.; Ahmed, M.; Iqbal, M. Synthesis, Characterization, and Antibacterial Activities of Novel Sulfonamides Derived through Condensation of Amino Group Containing Drugs, Amino Acids, and Their Analogs. BioMed Res. Int. 2015, 2015, 938486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camoutsis, C.; Geronikaki, A.; Ciric, A.; Soković, M.; Zoumpoulakis, P.; Zervou, M. Sulfonamide-1,2,4-thiadiazole Derivatives as Antifungal and Antibacterial Agents: Synthesis, Biological Evaluation, Lipophilicity, and Conformational Studies. Chem. Pharm. Bull. 2010, 58, 160–167. [Google Scholar] [CrossRef] [Green Version]
- Qadir, M.A.; Ahmed, M.; Aslam, H.; Waseem, S.; Shafiq, M.I. Amidine Sulfonamides and Benzene Sulfonamides: Synthesis and Their Biological Evaluation. J. Chem. 2015, 2015, 524056. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.-D.; Liu, P.; Yang, Y.; Gao, F. Sulfonamide-1,3,5-triazine–thiazoles: Discovery of a novel class of antidiabetic agents via inhibition of DPP-4. RSC Adv. 2016, 6, 83438–83447. [Google Scholar] [CrossRef]
- Durgapal, S.D.; Soman, S.S. Evaluation of novel coumarin-proline sulfonamide hybrids as anticancer and antidiabetic agents. Synth. Commun. 2019, 49, 1–15. [Google Scholar] [CrossRef]
- Badgujar, J.R.; More, D.H.; Meshram, J.S. Synthesis, Antimicrobial and Antioxidant Activity of Pyrazole Based Sulfonamide Derivatives. Indian J. Microbiol. 2018, 58, 93–99. [Google Scholar] [CrossRef]
- Dragostin, O.M.; Samal, S.K.; Lupascu, F.; Pânzariu, A.; Dubruel, P.; Lupascu, D.; Tuchilus, C.; Vasile, C.; Profire, L. Development and Characterization of Novel Films Based on Sulfonamide-Chitosan Derivatives for Potential Wound Dressing. Int. J. Mol. Sci. 2015, 16, 29843–29855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bérubé, G. An overview of molecular hybrids in drug discovery. Expert Opin. Drug Discov. 2016, 11, 281–305. [Google Scholar] [CrossRef] [PubMed]
- Charitos, G.; Trafalis, D.T.; Dalezis, P.; Potamitis, C.; Sarli, V.; Zoumpoulakis, P.; Camoutsis, C. Synthesis and anticancer activity of novel 3,6-disubstituted 1,2,4-triazolo-[3,4-b]-1,3,4-thiadiazole derivatives. Arab. J. Chem. 2019, 12, 4784–4794. [Google Scholar] [CrossRef] [Green Version]
- OpenTox. Available online: http://www.opentox.org/toxicity-prediction (accessed on 5 May 2018).
- ToxPredict. Available online: https://apps.ideaconsult.net/ToxPredict (accessed on 11 May 2018).
- PROTOX. Available online: http://tox.charite.de/tox (accessed on 11 May 2018).
- Miyagawa, M. Globally harmonized system of classification and labelling of chemicals (GHS) and its implementation in Japan. Nihon Eiseigaku Zasshi 2010, 65, 5–13. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Geronikaki, A.; Fesatidou, M.; Kartsev, V.; Macaev, F. Synthesis and biological evaluation of potent antifungal agents. Curr. Top. Med. Chem. 2013, 13, 2684–2733. [Google Scholar] [CrossRef]
- Benson, T.; Walsh, C.T.; Massey, V. Kinetic Characterization of Wild-Type and S229A Mutant MurB: Evidence for the Role of Ser 229 as a General Acid. Biochemistry 1997, 36, 796–805. [Google Scholar] [CrossRef]
- Yang, Y.; Severin, A.; Chopra, R.; Krishnamurthy, G.; Singh, G.; Hu, W.; Keeney, D.; Svenson, K.; Petersen, P.J.; Labthavikul, P.; et al. 3,5-dioxopyrazolidines, novel inhibitors of UDP-N- acetylenolpyruvylglucosamine reductase (MurB) with activity against gram-positive bacteria. Antimicrob. Agents Chemother. 2006, 50, 556–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haroun, M.; Tratrat, C.; Kolokotroni, A.; Petrou, A.; Geronikaki, A.; Ivanov, M.; Kostic, M.; Sokovic, M.; Carazo, A.; Mladěnka, P.; et al. 5-Benzyliden-2-(5-methylthiazol-2-ylimino)thiazolidin-4-ones as Antimicrobial Agents. Design, Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics 2021, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Cortellis Drug Discovery Intelligence. Available online: https://www.cortellis.com/drugdiscovery/ (accessed on 7 May 2021).
- Trafalis, D.; Galenica, S.A. New 1,2,4-triazolo-[3,4-b]-1,3,4-thiadiazole Derivatives. Patent No. WO 2018011414, 14 July 2016. [Google Scholar]
- Bajusz, D.; Rácz, A.; Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminformatics 2015, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Jasial, S.; Hu, Y.; Vogt, M.; Bajorath, J. Activity-relevant similarity values for fingerprints and implications for similarity searching. F1000Research 2016, 5, 591. [Google Scholar] [CrossRef] [Green Version]
- Kartsev, V.; Lichitsky, B.; Geronikaki, A.; Petrou, A.; Smiljkovic, M.; Kostic, M.; Ranadovic, O.; Soković, M. Design, synthesis and antimicrobial activity of usnic acid derivatives. MedChemComm 2018, 9, 870–882. [Google Scholar] [CrossRef] [Green Version]
- Kostić, M.; Smiljković, M.; Petrović, J.; Glamočilija, J.; Barros, L.; Ferreira, I.C.F.R.; Ćirić, A.; Soković, M. Chemical, nutritive composition and a wide range of bioactive properties of honey mushroom Armillariamellea (Vahl: Fr.) Kummer. Food Funct. 2017, 8, 3239–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cady, N.C.; Mckean, K.A.; Behnke, J.; Kubec, R.; Mosier, A.P.; Kasper, S.H.; Burz, D.S.; Musah, R.A. Inhibition of Biofilm Formation, Quorum Sensing and Infection in Pseudomonas aeruginosa by Natural Products-Inspired Organosulfur Compounds. PLoS ONE 2012, 7, e38492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kritsi, E.; Matsoukas, M.-T.; Potamitis, C.; Detsi, A.; Ivanov, M.; Sokovic, M.; Zoumpoulakis, P. Novel Hit Compounds as Putative Antifungals: The Case of Aspergillus fumigatus. Molecules 2019, 24, 3853. [Google Scholar] [CrossRef] [Green Version]
- Aleksić, M.; Stanisavljević, D.; Smiljković, M.; Vasiljević, P.; Stevanovic, M.; Sokovic, M.; Stojković, D. Pyrimethanil: Between efficient fungicide against Aspergillus rot on cherry tomato and cytotoxic agent on human cell lines. Ann. Appl. Biol. 2019, 175, 228–235. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]






| Compounds Number | Structure | Compounds | Structure |
|---|---|---|---|
| 1 |  | 10 |  |
| 2 |  | 11 |  |
| 3 |  | 12 |  |
| 4 |  | 13 |  |
| 5 |  | 14 |  |
| 6 |  | 15 |  |
| 7 |  | 16 |  |
| 8 |  | 17 |  |
| 9 |  | 18 |  |
| 19 |  |
| Compounds | B.c | S.a | L.m. | P.a. | E. coli | S.t | |
|---|---|---|---|---|---|---|---|
| 1 | MIC | 20 ± 0.000 | 15 ± 0.004 | 15 ± 0.004 | 150 ± 0.04 | 5 ± 0.000 | 15 ± 0.002 |
| MBC | 40 ± 0.000 | 40 ± 0.000 | 20 ± 0.008 | 200 ± 0.070 | 10 ± 0.00 | 20 ± 0.000 | |
| 2 | MIC | 15 ± 0.002 | 10 ± 0.000 | 15 ± 0.004 | 20 ± 0.000 | 5 ± 0.000 | 15 ± 0.004 |
| MBC | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 | 20 ± 0.004 | |
| 3 | MIC | 15 ± 0.004 | 30 ± 0.007 | 5 ± 0.000 | 15 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.004 | 40 ± 0.006 | 10 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | |
| 4 | MIC | 15 ± 0.002 | 10 ± 0.000 | 15 ± 0.000 | 30 ± 0.008 | 10 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.004 | 0.20 ± 0.002 | 0.020 ± 0.002 | 0.040 ± 0.000 | 0.020 ± 0.000 | 0.020 ± 0.000 | |
| 5 | MIC | 15 ± 0.002 | 20 ± 0.000 | 30 ± 0.000 | 15 ± 0.004 | 5 ± 0.000 | 3 ± 0.005 |
| MBC | 20 ± 0.002 | 36 ± 0.004 | 40 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 40 ± 0.000 | |
| 6 | MIC | 10 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 | 20 ± 0.000 | 8 ± 0.000 | 5 ± 0.000 |
| MBC | 20 ± 0.000 | 20 ± 0.000 | 36 ± 0.004 | 40 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | |
| 7 | MIC | 15 ± 0.002 | 20 ± 0.000 | 15 ± 0.002 | 15 ± 0.004 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | |
| 8 | MIC | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 |
| MBC | 36 ± 0.005 | 40 ± 0.000 | 40 ± 0.000 | 73 ± 0.009 | 20 ± 0.000 | 20 ± 0.000 | |
| 9 | MIC | 20 ± 0.000 | 30 ± 0.000 | 10 ± 0.000 | 23 ± 0.004 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 40 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | |
| 10 | MIC | 15 ± 0.002 | 10 ± 0.000 | 23 ± 0.004 | 40 ± 0.000 | 8 ± 0.000 | 8 ± 0.000 |
| MBC | 20 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | 60 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | |
| 11 | MIC | 10 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 40 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 23 ± 0.004 | 40 ± 0.000 | 20 ± 0.000 | 67 ± 0.009 | 10 ± 0.000 | 20 ± 0.000 | |
| 12 | MIC | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 80 ± 0.000 | 5 ± 0.000 | 8 ± 0.000 |
| MBC | 40 ± 0.000 | 37 ± 0.005 | 20 ± 0.000 | 150 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | |
| 13 | MIC | 10 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 150 ± 0.020 | 3 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 200 ± 0.000 | 47 ± 0.009 | 20 ± 0.000 | |
| 14 | MIC | 15 ± 0.002 | 40 ± 0.000 | 40 ± 0.000 | 40 ± 0.000 | 8 ± 0.000 | 40 ± 0.000 |
| MBC | 20 ± 0.000 | 60 ± 0.000 | 80 ± 0.000 | 80 ± 0.000 | 13 ± 0.002 | 20 ± 0.000 | |
| 15 | MIC | 5 ± 0.000 | 20 ± 0.000 | 15 ± 0.002 | 36 ± 0.004 | 8 ± 0.000 | 15 ± 0.000 |
| MBC | 10 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 60 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | |
| 16 | MIC | 15 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 | 67 ± 0.009 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.000 | 80 ± 0.000 | 20 ± 0.000 | 80 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | |
| 17 | MIC | 8 ± 0.000 | 15 ± 0.004 | 10 ± 0.000 | 40 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 80 ± 0.000 | 10 ± 0.000 | 20 ± 0.007 | |
| 18 | MIC | 15 ± 0.002 | 30 ± 0.007 | 10 ± 0.000 | 37 ± 0.005 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 20 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 60 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | |
| 19 | MIC | 5 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 |
| MBC | 10 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | |
| Streptomyci | MIC | 25 ± 0.000 | 100 ± 0.000 | 150 ± 0.000 | 100 ± 0.000 | 100 ± 0.000 | 100 ± 0.000 |
| MBC | 50 ± 0.000 | 200 ± 0.010 | 300 ± 0.010 | 200 ± 0.010 | 200 ± 0.000 | 200 ± 0.010 | |
| Ampicillin | MIC | 100 ± 0.000 | 100 ± 0.000 | 150 ± 0.000 | 300 ± 0.010 | 150 ± 0.000 | 100 ± 0.000 |
| MBC | 150 ± 0.000 | 150 ± 0.000 | 300 ± 0.020 | 500 ± 0.010 | 200 ± 0.010 | 200 ± 0.000 |
| Compounds | MRSA | P.a. | E. coli | Compounds | MRSA | P.a. | E. coli | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | MIC | 20 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 11 | MIC | 10 ± 0.000 | 8 ± 0.000 | 10 ± 0.000 |
| MBC | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | MBC | 20 ± 0.007 | 10 ± 0.000 | 20 ± 0.000 | ||
| 2 | MIC | 30 ± 0.007 | 10 ± 0.000 | 10 ± 0.000 | 12 | MIC | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 |
| MBC | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | MBC | 40 ± 0.000 | 20 ± 0.000 | 37 ± 0.005 | ||
| 3 | MIC | 30 ± 0.007 | 15 ± 0.002 | 15 ± 0.002 | 13 | MIC | 30 ± 0.007 | 10 ± 0.000 | 20 ± 0.000 |
| MBC | 40 ± 0.000 | 40 ± 0.000 | 40 ± 0.000 | MBC | 40 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | ||
| 4 | MIC | 30 ± 0.007 | 10 ± 0.000 | 10 ± 0.000 | 14 | MIC | 67 ± 0.009 | 40 ± 0.000 | 30 ± 0.007 |
| MBC | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | MBC | 80 ± 0.000 | 80 ± 0.000 | 40 ± 0.000 | ||
| 5 | MIC | 20 ± 0.004 | 20 ± 0.004 | 20 ± 0.000 | 15 | MIC | 15 ± 0.002 | 80 ± 0.000 | 10 ± 0.000 |
| MBC | 40 ± 0.000 | 40 ± 0.000 | 40 ± 0.000 | MBC | 20 ± 0.000 | 150 ± 0.002 | 20 ± 0.000 | ||
| 6 | MIC | 20 ± 0.0002 | 15 ± 0.002 | 15 ± 0.002 | 16 | MIC | 20 ± 0.000 | 2 ± 0.000 | 15 ± 0.002 |
| MBC | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | MBC | 40 ± 0.000 | 5 ± 0.000 | 20 ± 0.000 | ||
| 7 | MIC | 20 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 17 | MIC | 15 ± 0.002 | 5 ± 0.000 | 10 ± 0.000 |
| MBC | 40 ± 0.000 | 20 ± 0.001 | 20 ± 0.000 | MBC | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | ||
| 8 | MIC | 80 ± 0.002 | 10 ± 0.000 | 10 ± 0.000 | 18 | MIC | 20 ± 0.000 | 40 ± 0.000 | 10 ± 0.000 |
| MBC | 150 ± 0.010 | 20 ± 0.000 | 20 ± 0.008 | MBC | 40 ± 0.000 | 73 ± 0.009 | 20 ± 0.000 | ||
| 9 | MIC | 30 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 19 | MIC | 20 ± 0.000 | 5 ± 0.000 | 15 ± 0.000 |
| MBC | 40 ± 0.000 | 20 ± 0.008 | 20 ± 0.000 | MBC | 40 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | ||
| 10 | MIC | 10 ± 0.001 | 15 ± 0.005 | 15 ± 0.004 | Streptomycin | MIC | 100 ± 0.000 | 50 ± 0.000 | 100 ± 0.000 |
| MBC | 20 ± 0.000 | 20 ± 0.008 | 20 ± 0.000 | MBC | - | 100 ± 0.000 | 200 ± 0.010 | ||
| Ampicillin | MIC | - | 200 ± 0.010 | 200 ± 0.010 | |||||
| MBC | - | - | - | ||||||
| Compound | MIC | 0.5 MIC |
|---|---|---|
| Biofilm inhibition (% compared to no treatment) | ||
| 2 | 49.82 ± 2.35 | 40.74 ± 8.89 |
| 3 | 75.10 ± 6.89 | 62.82 ± 4.56 |
| 6 | 50.85 ± 8.82 | 31.90 ± 6.98 |
| 7 | 44.13 ± 3.56 | 38.75 ± 2.11 |
| 19 | 47.84 ± 2.36 | 41.50 ± 1.08 |
| Ampicillin | 70.00 ± 10.23 | 52.36 ± 3.67 |
| Streptomycin | 63.56 ± 8.28 | 29.12 ± 1.22 |
| Compounds | A.f | A.v. | A.n. | T.v. | P.f. | P.v.c. | |
|---|---|---|---|---|---|---|---|
| 1 | MIC | 5 ± 0.000 | 2 ± 0.000 | 10 ± 0.000 | 2 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 |
| MFC | 10 ± 0.000 | 5 ± 0.000 | 20 ± 0.000 | 5 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | |
| 2 | MIC | 10 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 | 2 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 |
| MFC | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | 5 ± 0.000 | 20 ± 0.007 | 20 ± 0.000 | |
| 3 | MIC | 20 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 | 2 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 |
| MFC | 40 ± 0.000 | 20 ± 0.000 | 20 ± 0.002 | 5 ± 0.000 | 36 ± 0.004 | 36 ± 0.004 | |
| 4 | MIC | 2 ± 0.000 | 2 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 |
| MFC | 5 ± 0.000 | 5 ± 0.000 | 1 ± 0.000 | 1 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | |
| 5 | MIC | 20 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 | 8 ± 0.000 | 33 ± 0.005 | 20 ± 0.000 |
| MFC | 36 ± 0.007 | 20 ± 0.070 | 20 ± 0.000 | 10 ± 0.000 | 40 ± 0.000 | 40 ± 0.000 | |
| 6 | MIC | 5 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | 2 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 |
| MFC | 10 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | |
| 7 | MIC | 10 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | 20 ± 0.000 |
| MFC | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 36 ± 0.004 | |
| 8 | MIC | 10 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 | 5 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 |
| MFC | 20 ± 0.007 | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | |
| 9 | MIC | 5 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 36 ± 0.005 | 30 ± 0.007 |
| MFC | 10 ± 0.000 | 10 ± 0.000 | 20 ± 0.002 | 10 ± 0.000 | 80 ± 0.000 | 40 ± 0.000 | |
| 10 | MIC | 32 ± 0.004 | 20 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 |
| MFC | 40 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | |
| 11 | MIC | 20 ± 0.000 | 5 ± 0.000 | 20 ± 0.000 | 2 ± 0.001 | 2 ± 0.000 | 2 ± 0.000 |
| MFC | 40 ± 0.000 | 10 ± 0.000 | 32 ± 0.005 | 5 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | |
| 12 | MIC | 5 ± 0.000 | 10 ± 0.000 | 15 ± 0.002 | 5 ± 0.000 | 40 ± 0.000 | 20 ± 0.000 |
| MFC | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.003 | 67 ± 0.009 | 40 ± 0.000 | |
| 13 | MIC | 40 ± 0.000 | 20 ± 0.007 | 20 ± 0.000 | 2 ± 0.000 | 40 ± 0.000 | 32 ± 0.005 |
| MFC | 67 ± 0.009 | 36 ± 0.005 | 40 ± 0.000 | 5 ± 0.000 | 80 ± 0.000 | 40 ± 0.000 | |
| 14 | MIC | 10 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 30 ± 0.007 | 32 ± 0.005 |
| MFC | 36 ± 0.007 | 20 ± 0.000 | 20 ± 0.001 | 10 ± 0.003 | 40 ± 0.000 | 40 ± 0.000 | |
| 15 | MIC | 15 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 | 5 ± 0.000 | 30 ± 0.007 | 30 ± 0.007 |
| MFC | 20 ± 0.000 | 20 ± 0.000 | 37 ± 0.005 | 10 ± 0.000 | 40 ± 0.000 | 36 ± 0.005 | |
| 16 | MIC | 20 ± 0.000 | 10 ± 0.000 | 5 ± 0.000 | 5 ± 0.000 | 10 ± 0.000 | 20 ± 0.000 |
| MFC | 40 ± 0.000 | 20 ± 0.000 | 1 ± 0.000 | 8 ± 0.000 | 20 ± 0.000 | 40 ± 0.000 | |
| 17 | MIC | 20 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 | 5 ± 0.000 | 30 ± 0.007 | 30 ± 0.007 |
| MFC | 40 ± 0.000 | 40 ± 0.000 | 32 ± 0.005 | 10 ± 0.003 | 40 ± 0.000 | 40 ± 0.000 | |
| 18 | MIC | 20 ± 0.000 | 10 ± 0.000 | 10 ± 0.000 | 8 ± 0.000 | 20 ± 0.000 | 30 ± 0.007 |
| MFC | 32 ± 0.004 | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 40 ± 0.000 | 36 ± 0.005 | |
| 19 | MIC | 20 ± 0.000 | 20 ± 0.000 | 10 ± 0.000 | 8 ± 0.000 | 20 ± 0.000 | 20 ± 0.000 |
| MFC | 40 ± 0.000 | 36 ± 0.005 | 20 ± 0.000 | 10 ± 0.000 | 40 ± 0.000 | 40 ± 0.000 | |
| Ketoconazole | MIC | 20 ± 0.010 | 200 ± 0.000 | 200 ± 0.010 | 1000 ± 0.010 | 200 ± 0.000 | 200 ± 0.010 |
| MFC | 500 ± 0.030 | 500 ± 0.020 | 500 ± 0.030 | 1500 ± 0.020 | 500 ± 0.030 | 300 ± 0.010 | |
| Bifonazole | MIC | 150 ± 0.000 | 100 ± 0.000 | 150 ± 0.000 | 150 ± 0.000 | 200 ± 0.010 | 100 ± 0.000 |
| MFC | 200 ± 0.000 | 200 ± 0.010 | 200 ± 0.000 | 200 ± 0.010 | 250 ± 0.010 | 200 ± 0.000 | |
| Comp. | Est. Binding Energy (kcal/mol) | I-H * E. coli MurB | Residues E. coli MurB | ||||
|---|---|---|---|---|---|---|---|
| E. coli Gyrase 1KZN | Thymidylate kinase 4QGG | E. coli Primase 1DDE | E. coli MurA JV4T | E. coli MurB 2Q85 | |||
| 1 | −4.45 | −3.12 | - | - | −7.15 | 2 | Arg158, Ser228 |
| 2 | −2.89 | - | - | −6.19 | −10.92 | 3 | Arg158, Ser228, Asn232 |
| 3 | −5.32 | −1.51 | −6.29 | −5.27 | −12.18 | 3 | Arg158, Arg213, Ser228 |
| 4 | −6.95 | −1.22 | −4.28 | −5.33 | −8.03 | 2 | Arg158, Ser228 |
| 5 | −2.39 | −1.18 | −2.44 | −6.01 | −9.21 | 2 | Arg213, Ser228 |
| 6 | −4.78 | - | −4.93 | −5.88 | −12.10 | 3 | Gly122, Arg213, Ser228 |
| 7 | −3.16 | - | −5.36 | −5.32 | −10.23 | 3 | Arg213, Ser228, Lys261 |
| 8 | - | −3.22 | - | −6.28 | −8.62 | 2 | Arg213, Ser228 |
| 9 | −4.77 | −2.13 | −3.95 | - | −10.07 | 3 | Arg213, Ser228, Asn232 |
| 10 | −4.01 | - | −2.55 | −2.53 | −10.11 | 3 | Arg213, Ser228 |
| 11 | −5.51 | −3.25 | - | - | −9.92 | 3 | Arg158, Arg213, Ser228 |
| 12 | −5.14 | −2.99 | - | - | −10.03 | 3 | Arg213, Ser228 |
| 13 | −3.12 | - | - | −4.83 | −7.75 | 2 | Arg213, Ser228 |
| 14 | −2.17 | - | - | −7.77 | 2 | Arg158, Ser228 | |
| 15 | - | - | −2.09 | −2.25 | −8.94 | 2 | Arg213, Ser228 |
| 16 | - | - | −3.85 | −6.94 | −8.13 | 2 | Ser228, Lys261 |
| 17 | −4.28 | −4.11 | - | −5.71 | −8.74 | 2 | Arg158, Ser228 |
| 18 | - | −3.15 | −4.16 | −3.82 | −9.71 | 2 | Arg213, Ser228 |
| 19 | −5.13 | −5.02 | −6.11 | −6.33 | −13.56 | 4 | Arg213, Ser228, Lys261 |
| Est. Binding Energy(kcal/mol) | |||||
|---|---|---|---|---|---|
| N/N | DNA TopoIV 1S16 | CYP51 of C. albicans 5V5Z | I-H | Residues CYP51 of C. albicans | Interactions with HEM601 |
| 1 | −4.16 | −7.96 | 1 | Tyr132 | Hydrophobic |
| 2 | −2.15 | −7.88 | 1 | Tyr64 | - |
| 3 | −3.19 | −7.63 | 1 | Tyr118 | - |
| 4 | −2.88 | −8.63 | - | - | aromatic |
| 5 | - | −7.55 | 1 | Tyr118 | Hydrophobic |
| 6 | −5.29 | −8.11 | 1 | Tyr64 | - |
| 7 | −3.36 | −8.07 | 1 | Tyr132 | - |
| 8 | - | −8.15 | 1 | Tyr118 | Hydrophobic |
| 9 | - | −7.07 | 1 | Tyr132 | - |
| 10 | −2.58 | −7.13 | 1 | Tyr132 | Hydrophobic |
| 11 | −3.95 | −7.03 | 1 | Tyr118 | - |
| 12 | - | −6.87 | 1 | Tyr118 | - |
| 13 | −3.77 | −6.96 | 1 | Tyr132 | - |
| 14 | −4.55 | −8.26 | 1 | Tyr132 | Hydrophobic |
| 15 | - | −8.22 | 1 | Met508 | Hydrophobic |
| 16 | −1.12 | −7.31 | 1 | Tyr132 | - |
| 17 | −3.74 | −7.59 | 1 | Tyr118 | - |
| 18 | −5.19 | −8.22 | 1 | Tyr132 | Hydrophobic |
| 19 | −1.52 | −7.91 | - | - | Hydrophobic, aromatic |
| ketoconazole | - | −8.23 | 1 | Tyr64 | Hydrophobic, aromatic |
| No | MW | Number of HBA a | Number of HBD b | Log Po/w (iLOGP) c | Log S d | TPSA e | BBB Permeant f | Lipinski, Ghose, Veber, Egan, and Muegge Violations | Bioavailability Score | Drug-Likeness Model Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 503.59 | 9 | 0 | 3.31 | Moderately soluble | 144.77 | No | 1 | 0.55 | 0.56 |
| 2 | 533.62 | 10 | 0 | 3.23 | Poorly soluble | 154.00 | No | 2 | 0.17 | 0.26 |
| 3 | 485.58 | 8 | 0 | 3.09 | Poorly soluble | 135.54 | No | 0 | 0.55 | 0.53 |
| 4 | 517.62 | 9 | 0 | 3.37 | Poorly soluble | 144.77 | No | 1 | 0.55 | 0.45 |
| 5 | 487.60 | 8 | 0 | 3.36 | Poorly soluble | 135.54 | No | 0 | 0.55 | 0.71 |
| 6 | 489.57 | 9 | 0 | 3.54 | Poorly soluble | 144.77 | No | 0 | 0.55 | 0.49 |
| 7 | 474.56 | 8 | 1 | 2.98 | Moderately soluble | 161.56 | No | 0 | 0.55 | 0.32 |
| 8 | 503.59 | 9 | 0 | 2.55 | Poorly soluble | 144.77 | No | 1 | 0.55 | 0.42 |
| 9 | 563.65 | 11 | 0 | 3.96 | Poorly soluble | 163.22 | No | 2 | 0.17 | 0.14 |
| 10 | 518.57 | 10 | 0 | 3.30 | Moderately soluble | 181.36 | No | 2 | 0.17 | −0.34 |
| 11 | 538.98 | 10 | 0 | 2.91 | Poorly soluble | 181.36 | No | 2 | 0.17 | −0.10 |
| 12 | 473.57 | 8 | 0 | 3.18 | Poorly soluble | 135.54 | No | 0 | 0.55 | 0.56 |
| 13 | 493.99 | 8 | 0 | 3.48 | Poorly soluble | 135.54 | No | 0 | 0.55 | 0.94 |
| 14 | 460.53 | 9 | 0 | 2.72 | Moderately soluble | 148.46 | No | 0 | 0.55 | 0.89 |
| 15 | 460.53 | 9 | 0 | 2.92 | Moderately soluble | 148.43 | No | 0 | 0.55 | 0.67 |
| 16 | 539.43 | 9 | 0 | 2.80 | Poorly soluble | 148.43 | No | 1 | 0.55 | 0.21 |
| 17 | 474.56 | 8 | 1 | 2.85 | Moderately soluble | 161.56 | No | 0 | 0.55 | 0.43 |
| 18 | 474.56 | 8 | 1 | 2.78 | Moderately soluble | 161.56 | No | 0 | 0.55 | 0.76 |
| 19 | 459.54 | 8 | 0 | 3.14 | Poorly soluble | 135.54 | No | 0 | 0.55 | 0.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamoutsis, C.; Fesatidou, M.; Petrou, A.; Geronikaki, A.; Poroikov, V.; Ivanov, M.; Soković, M.; Ćirić, A.; Carazo, A.; Mladěnka, P. Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics 2021, 10, 804. https://doi.org/10.3390/antibiotics10070804
Kamoutsis C, Fesatidou M, Petrou A, Geronikaki A, Poroikov V, Ivanov M, Soković M, Ćirić A, Carazo A, Mladěnka P. Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics. 2021; 10(7):804. https://doi.org/10.3390/antibiotics10070804
Chicago/Turabian StyleKamoutsis, Charalampos, Maria Fesatidou, Anthi Petrou, Athina Geronikaki, Vladimir Poroikov, Marija Ivanov, Marina Soković, Ana Ćirić, Alejandro Carazo, and Přemysl Mladěnka. 2021. "Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies" Antibiotics 10, no. 7: 804. https://doi.org/10.3390/antibiotics10070804
APA StyleKamoutsis, C., Fesatidou, M., Petrou, A., Geronikaki, A., Poroikov, V., Ivanov, M., Soković, M., Ćirić, A., Carazo, A., & Mladěnka, P. (2021). Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics, 10(7), 804. https://doi.org/10.3390/antibiotics10070804