Quantification of Total and Viable Cells and Determination of Serogroups and Antibiotic Resistance Patterns of Listeria monocytogenes in Chicken Meat from the North-Western Iberian Peninsula
Abstract
:1. Introduction
2. Results
2.1. Levels of Microorganisms Indicating Quality of Hygiene
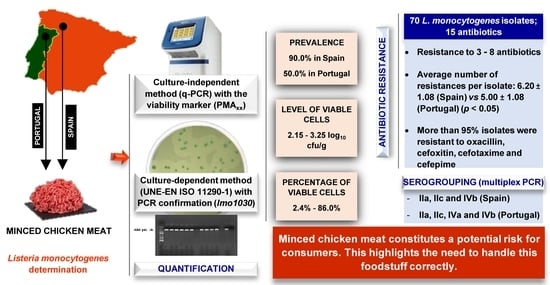
2.2. Prevalence and Levels of Listeria monocytogenes
2.3. Serogroups of Listeria monocytogenes
2.4. Susceptibility to Antibiotics of Listeria monocytogenes
3. Discussion
3.1. Levels of Microorganisms Indicating the Quality of Hygiene
3.2. Prevalence and Levels of Listeria monocytogenes
3.3. Serogroups of Listeria monocytogenes
3.4. Susceptibility of Antibiotics
4. Material and Methods
4.1. Sample Collection
4.2. Counts of Microorganisms Indicating the Quality of Hygiene
4.3. Isolation and Identification of Listeria monocytogenes
4.4. Detecion and Quantification of Listeria monocytogenes by q-PCR
4.5. Multiplex PCR Serogrouping of Listeria monocytogenes Isolates
4.6. Antibiotic Susceptibility Tests
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Consumo Carne de Pollo. Available online: http://www.fao.org/faostat/es/#data/QL (accessed on 22 September 2021).
- Cardoso Pereira, P.M.C.; Baltazar Vicente, A.F.R. Meat nutritional composition and nutritive role in the human diet. Meat Sci. 2013, 93, 586–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capita, R.; Castaño-Arriba, A.; Rodríguez-Melcón, C.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C. Diversity, antibiotic resistance, and biofilm-forming ability of enterobacteria isolated from red meat and poultry preparations. Microorganisms 2020, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Microbial loads and antibiotic resistance patterns of Staphylococcus aureus in different types of raw poultry-based meat preparations. Poult. Sci. 2017, 96, 4046–4052. [Google Scholar] [CrossRef] [PubMed]
- Selvan, P.; Naremdra, B.R.; Sureshkumar, S.; Vankataramanujam, V. Microbial quality of retail meat products available in Chennai city. Am. J. Food Technol. 2007, 2, 55–59. [Google Scholar]
- Del Río, E.; Panizo-Morán, M.; Prieto, M.; Alonso-Calleja, C.; Capita, R. Effect of various chemical decontamination treatments on natural microflora and sensory characteristics of poultry. Int. J. Food Microbiol. 2007, 115, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Lactic acid concentrations that reduce microbial load yet minimally impact colour and sensory characteristics of beef. Meat Sci. 2017, 129, 169–175. [Google Scholar] [CrossRef]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; Carballo, J.; García-Fernández, C.; Capita, R.; Alonso-Calleja, C. Structure and viability of 24- and 72-h-old biofilms formed by four pathogenic bacteria on polystyrene and glass contact surfaces. Food Microbiol. 2018, 76, 513–517. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Susceptibility of Listeria monocytogenes planktonic cultures and biofilms to sodium hypochlorite and benzalkonium chloride. Food Microbiol. 2019, 82, 533–540. [Google Scholar] [CrossRef]
- Maertens de Noordhout, C.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- EFSA; ECDC. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar]
- Muchaamba, F.; Eshwar, A.K.; Stevens, M.J.A.; Stephan, R.; Tasara, T. Different shades of Listeria monocytogenes: Strain, serotype, and lineage-based variability in virulence and stress tolerance profiles. Front. Microbiol. 2022, 4, 792162. [Google Scholar] [CrossRef] [PubMed]
- Tîrzu, E.; Herwman, V.; Nichita, I.; Morar, A.; Imre, M.; Ban-Cucerzan, A.; Bucur, I.; Tîrziu, A.; Mateiu-Petrec, O.C.; Imre, K. Diversity and antibiotic resistance profiles of Listeria monocytogenes serogroups in different food products from the Transylvania Region of Central Romania. J. Food Prot. 2022, 85, 54–59. [Google Scholar] [CrossRef]
- Alonso-Calleja, C.; Gómez-Fernández, S.; Carballo, J.; Capita, R. Prevalence, molecular typing, and determination of the biofilm-forming ability of Listeria monocytogenes serotypes from poultry meat and poultry preparations in Spain. Microorganisms 2019, 7, 529. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Felices-Mercado, A.; García-Fernández, C.; Alonso-Calleja, C. Characterization of Listeria monocytogenes originating from the Spanish meat-processing chain. Foods 2019, 8, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barretta, C.; Verruck, S.; Maran, M.B.; Mauricio, L.S.; Mirotto, L.; Vieira, C.R.W.; Prudencio, E.S. Listeria monocytogenes survival in raw Atlantic salmon (Salmo salar) fillet under in vitro simulated gastrointestinal conditions by culture, qPCR and PMA-qPCR detection methods. LWT 2019, 107, 132–137. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 14 November 2022).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 22 September 2021).
- OCDE. Antimicrobial Resistance. Tackling the Burden in the European Union. 2019. Available online: https://www.oecd.org/health/health-systems/AMR-Tackling-the-Burden-in-the-EU-OECD-ECDC-Briefing-Note-2019.pdf (accessed on 22 September 2021).
- González-Gutiérrez, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Microbial load and antibiotic resistance in raw beef preparations from northwest Spain. Food Sci. Nutr. 2019, 8, 777–785. [Google Scholar] [CrossRef]
- ICMSF. Microorganisms in Foods 8. Use of Data for Assessing Process Control and Product Acceptance; Springer: New York, NY, USA, 2011. [Google Scholar]
- IFST. Development and use of microbiological criteria for foods. Food Sci. Technol. Today 1997, 11, 137–176. [Google Scholar]
- OJEU. Commission regulation (EC) N° 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2005, L338, 1–26. [Google Scholar]
- Lerasle, M.; Federighi, M.; Simonin, H.; Anthoine, V.; Rezé, S.; Chéret, R.; Guillou, S. Combined use of modified atmosphere packaging and high pressure to extend the shelf-life of raw poultry sausage. Innov. Food Sci. Emerg. Technol. 2014, 23, 54–60. [Google Scholar] [CrossRef]
- Łaszkiewicz, B.; Szymański, P.; Kołożyn-Krajewska, D. The effect of selected lactic acid bacterial strains on the technological and microbiological quality of mechanically separated poultry meat cured with a reduced amount of sodium nitrite. Poult. Sci. 2021, 100, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Calleja, C.; Martínez-Fernández, B.; Prieto, M.; Capita, R. Microbiological quality of vacuum-packed retail ostrich meat in Spain. Food Microbiol. 2004, 21, 241–246. [Google Scholar] [CrossRef]
- Gashe, B.A.; Mpuchane, S. Prevalence of salmonellae on beef products at butcheries and their antibiotic resistance profiles. J. Food Sci. 2000, 65, 880–883. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Occurrence of Listeria species in retail poultry meat and comparison of a cultural/immunoassay for their detection. Int. J. Food Microbiol. 2001, 65, 75–82. [Google Scholar] [CrossRef]
- Pascual-Anderson, M.R. Microbiología Alimentaria. Metodología Analítica Para Alimentos Y Bebidas; Diaz de Santos: Madrid, Spain, 1992. [Google Scholar]
- Castaño-Arriba, A.; González-Machado, C.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Antibiotic resistance and biofilm-forming ability in enterococcal isolates from red meat and poultry preparations. Pathogens 2020, 9, 1021. [Google Scholar] [CrossRef]
- Dainty, R.H.; Mackey, B.M. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Bacteriol. Symp. Suppl. 1992, 73, 103S–114S. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C.; García-Arias, M.T.; Moreno, B.; García-Fernández, M.C. Methods to detect the occurrence of various indicator bacteria on the surface of retail poultry in Spain. J. Food Sci. 2002, 67, 765–771. [Google Scholar] [CrossRef]
- Jay, J.M. A Review of aerobic and psychrotrophic plate count procedures for fresh meat and poultry products. J. Food Prot. 2002, 65, 1200–1206. [Google Scholar] [CrossRef]
- Andritsos, N.D.; Mataragas, M.; Mavrou, E.; Stamatiou, A.; Drosinos, E.H. The microbiological condition of minced pork prepared at retail stores in Athens, Greece. Meat Sci. 2012, 91, 480–489. [Google Scholar] [CrossRef]
- Wong, T.; Whyte, R.J.; Cornelius, A.J.; Hudson, J.A. Enumeration of Campylobacter and Salmonella on chicken packs. Br. Food J. 2004, 106, 651–662. [Google Scholar] [CrossRef]
- Fraqueza, M.J.; Ferreira, M.C.; Barreto, A.S. Spoilage of light (PSE-like) and dark turkey meat under aerobic or modified atmosphere package: Microbial indicators and their relationship with total volatile basic nitrogen. Br. Poult. Sci. 2008, 49, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Säde, E.; Murros, A.; Björkroth, J. Predominant enterobacteria on modified-atmosphere packaged meat and poultry. Food Microbiol. 2013, 34, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Burbano, E.M.; Carrascal, A.K.; Mercado, M.; Poutou, R. Validación de PCR para Listeria monocytogenes en Leches. Aliment. Hoy 2011, 10, 1–10. [Google Scholar]
- De la Rosa Zariñana, A.E.; Crosby-Galván, M.M.; Ramírez-Guzmán, M.E.; Hernández-Sánchez, D.; Mata-Espinoza, M.A. Standardization of PCR technique for detecting Listeria monocytogenes in chicken, beef and pork. Ecosistemas y Recur. Agropecu. 2018, 5, 25–34. [Google Scholar] [CrossRef]
- Poutou, R.M.; Burbano, S.; Sierra, K.; Torres, A.; Carrascal, K.; Mercado, M. Estandarización de la extracción de ADN y validación de la PCR múltiple para detectar Listeria monocytogenes en queso, leche, carne de res y pollo. Univ. Sci. 2005, 10, 61–78. [Google Scholar]
- Jamshidi, A.; Zeinali, T. Significance and characteristics of Listeria monocytogenes in poultry products. Int. J. Food Sci. 2019, 2019, 7835253. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Hernando, A.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control. 2012, 23, 37–41. [Google Scholar] [CrossRef]
- Oliveira, T.S.; Varjão, L.M.; da Silva, L.N.N.; Pereira, R.C.L.; Hofer, E.; Vallim, D.C.; de Castro Almeida, R.C. Listeria monocytogenes at chicken slaughterhouse: Occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility. Food Control. 2012, 88, 131–138. [Google Scholar] [CrossRef]
- Gonçalves-Tenório, A.; Nunes Silva, B.; Rodrigues, V.; Cadavez, V.; Gonzales-Barron, U. Prevalence of pathogens in poultry meat: A meta-analysis of European published surveys. Foods 2018, 7, 69. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Réu, C.; Sousa, J.C.; Pestana, N.; Peixe, L. Incidence of susceptibility to antimicrobial agents of Listeria spp. and Listeria monocytogenes isolated from poultry carcasses in Porto, Portugal. J. Food Prot. 2002, 65, 1888–1893. [Google Scholar] [CrossRef] [PubMed]
- Mena, C.; Almeida, G.; Carneiro, L.; Teixeira, P.; Hogg, T.; Gibbs, P.A. Incidence of Listeria monocytogenes in different food products commercialized in Portugal. Food Microbiol. 2004, 21, 213–216. [Google Scholar] [CrossRef]
- Sugiri, Y.D.; Gölz, G.; Meeyam, T.; Baumann, M.P.O.; Kleer, J.; Chaisowwong, W.; Alter, T. Prevalence and antimicrobial susceptibility of Listeria monocytogenes on chicken carcasses in Bandung, Indonesia. J. Food Prot. 2014, 77, 1407–1410. [Google Scholar] [CrossRef] [PubMed]
- Rørvik, L.M.; Yndestad, M. Listeria monocytogenes in foods in Norway. Int. J. Food Microbiol. 1991, 13, 97–104. [Google Scholar] [CrossRef]
- Martín, B.; Jofré, A.; Garriga, M.; Hugas, M.; Aymerich, T. Quantification of Listeria monocytogenes in fermented sausages by MPN-PCR method. Lett. Appl. Microbiol. 2004, 39, 290–295. [Google Scholar] [CrossRef]
- Rantsiou, K.; Alessandria, V.; Urso, R.; Dolci, P.; Cocolin, M. Detection quantification and vitality of Listeria monocytogenes in food as determined by quantitative PCR. Int. J. Food Microbiol. 2008, 21, 99–105. [Google Scholar] [CrossRef]
- AESAN. Listeriosis. 2019. Available online: https://www.aesan.gob.es/AECOSAN/web/seguridad_alimentaria/subdetalle/listeria.htm (accessed on 21 September 2021).
- Rodríguez-Lázaro, D.; Jofré, A.; Aymerich, T.; Hugas, M.; Pla, M. Rapid quantitative detection of Listeria monocytogenes in meat products by Real-Time PCR. Appl. Environ. Microbiol. 2004, 70, 6299–6301. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Lázaro, D.; Jofré, A.; Aymerich, T.; Garriga, M.; Pla, M. Rapid quantitative detection of Listeria monocytogenes in salmon products: Evaluation of Pre–Real-Time PCR strategies. J. Food Prot. 2005, 68, 1467–1471. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, W.; Xu, H.; Aguilar, Z.P.; Nagendra, P.S.; Wei, H. Propidium monoazide combined with real-time PCR for selective detection of viable Staphylococcus aureus in milk powder and meat products. J. Dairy Sci. 2015, 98, 1625–1633. [Google Scholar] [CrossRef] [Green Version]
- Korsak, D.; Borek, A.; Daniluk, S.; Grabowska, A.; Pappelbaum, K. Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 2012, 158, 203–208. [Google Scholar] [CrossRef]
- Chen, M.; Cheng, J.; Zhang, J.; Chen, Y.; Zeng, H.; Xue, L.; Lei, T.; Pang, R.; Wu, S.; Wu, H.; et al. Isolation, potential virulence, and population diversity of Listeria monocytogenes from meat and meat products in China. Front. Microbiol. 2019, 10, 946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braga, V.; Vázquez, S.; Vico, V.; Pastorino, V.; Mota, M.I.; Legnani, M.; Schelotto, F.; Lancibidad, G.; Varela, G. Prevalence and serotype distribution of Listeria monocytogenes isolated from foods in Montevideo-Uruguay. Braz. J. Microbiol. 2017, 48, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Raschle, S.; Stephan, R.; Stevens, M.J.A.; Cernela, N.; Zurfuh, K.; Muchaamba, F.; Nüesch-Inderbinen, M. Environmental dissemination of pathogenic Listeria monocytogenes in fowing surface waters in Switzerland. Sci. Rep. 2021, 11, 9066. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, W.; Wang, J.; Xu, B.; Liu, H.; Dong, Q.; Zhang, X. 10-year molecular surveillance of Listeria monocytogenes using whole-genome sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 2020, 11, 551020. [Google Scholar] [CrossRef]
- Wu, S.; Wu, Q.; Zhang, J.; Chen, M.; Yan, Z.; Hu, H. Listeria monocytogenes prevalence and characteristics in retail foods in China. PLoS ONE 2015, 10, e0136682. [Google Scholar] [CrossRef]
- Lachtara, B.; Wieczorek, K.; Osek, J. Genetic diversity and relationships of Listeria monocytogenes serogroup IIa isolated in Poland. Microorganisms 2022, 10, 532. [Google Scholar] [CrossRef]
- Shimojima, Y.; Ida, M.; Nishino, Y.; Ishitsuka, R.; Kuroda, S.; Hirai, A.; Sadamasu, K.; Nakama, A.; Kai, A. Multiplex PCR serogrouping of Listeria monocytogenes isolated in Japan. J. Vet. Med. Sci. 2016, 78, 477–479. [Google Scholar] [CrossRef] [Green Version]
- Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Hernández, M.; Abad, D.; Rodríguez-Grande, J.; Ocampo-Sosa, A.A.; Martínez-Suárez, J.V. Prevalence and population diversity of Listeria monocytogenes isolated from dairy cattle farms in the Cantabria Region of Spain. Animals 2022, 12, 2477. [Google Scholar] [CrossRef]
- Amajoud, N.; Leclercq, A.; Soriano, J.M.; Bracq-Dieye, H.; El Maadoudi, M.; Senhaji, N.S.; Kounnoun, A.; Moura, A.; Lecuit, M.; Abrini, J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control. 2018, 84, 436–441. [Google Scholar] [CrossRef] [Green Version]
- Acciari, V.A.; Ruolo, A.; Torresi, M.; Ricci, L.; Pompei, A.; Marfoglia, C.; Valente, F.M.; Centorotola, G.; Conte, A.; Salini, R.; et al. Genetic diversity of Listeria monocytogenes strains contaminating food and food producing environment as single based sample in Italy (retrospective study). Int. J. Food Microbiol. 2022, 366, 109562. [Google Scholar] [CrossRef]
- Fallah, A.A.; Saei-Dehkordi, S.S.; Rahnama, M.; Tahmasby, H.; Mahzounieh, M. Prevalence and antimicrobial resistance patterns of Listeria species isolated from poultry products marketed in Iran. Food Control. 2012, 28, 327–332. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabusli, A.A.; Abu Goush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero, J.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Microbial load and antibiotic resistance patterns of Escherichia coli and Enterococcus faecalis isolates from the meat of wild and domestic pigeons. Foods 2019, 8, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019.
- OIE. OIE List of Antimicrobial Agents of Veterinary Importance. 2018. Available online: http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/Eng_OIE_List_antimicrobials_May2015.pdf (accessed on 23 September 2021).
- Adzitey, F.; Ali, G.R.R.; Huda, N.; Cogan, T.; Corry, J. Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from ducks, their rearing and processing environments in Penang, Malaysia. Food Control. 2013, 163, 607–614. [Google Scholar] [CrossRef]
- Bae, D.; Mezal, E.H.; Smiley, R.D.; Cheng, C.-M.; Khan, A.A. The sub-species characterization and antimicrobial resistance of Listeria monocytogenes isolated from domestic and imported food products from 2004 to 2011. Food Res. Int. 2014, 64, 656–663. [Google Scholar] [CrossRef]
- Doménech, E.; Jimenez-Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Control. 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Komora, N.; Bruschi, C.; Magalhães, R.; Ferreira, V.; Teixeira, P. Survival of Listeria monocytogenes with different antibiotic resistance patterns to food-associated stresses. Int. J. Food Microbiol. 2017, 245, 79–87. [Google Scholar] [CrossRef]
- Wang, X.-M.; Lü, X.-F.; Yin, L.; Liu, H.-F.; Zhang, W.-J.; Si, W.; Tu, S.-Y.; Shao, M.-L.; Liu, S.-G. Occurrence and antimicrobial susceptibility of Listeria monocytogenes isolates from retail raw foods. Food Control. 2013, 32, 153–158. [Google Scholar] [CrossRef]
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.; McAllister, A.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Doming, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Fernández, E.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Prevalence and antimicrobial resistance of Salmonella serotypes isolates from poultry in Spain: Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012, 153, 281–287. [Google Scholar] [CrossRef]
- Noll, M.; Kleta, S.; Al Dahouk, S. Antibiotic susceptibility of 259 Listeria monocytogenes strains isolated from food, food-processing plants and human samples in Germany. J. Infect. Public Health 2018, 11, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Cousin, M.A.; Jay, J.M.; Vasavada, P.C. Psychrotrophic microorganisms. In Compendium of Methods for the Microbiological Examination of Foods, 4th ed.; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 159–166. [Google Scholar]
- Baird, R.M.; Corry, J.E.J.; Curtis, G.D.W. Pharmacopeia of culture media for food microbiology. Int. J. Food Microbiol. 1987, 5, 221–222. [Google Scholar]
- Ryu, J.; Park, S.H.; Yeom, Y.S.; Shrivastav, A.; Lee, S.H.; Kim, Y.R.; Kim, H.Y. Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Control 2013, 32, 659–664. [Google Scholar] [CrossRef]
- Panera-Martínez, S.; Rodríguez-Melcón, C.; Serrano-Galán, V.; Alonso-Calleja, C.; Capita, R. Prevalence, quantification and antibiotic resistance of Listeria monocytogenes in poultry preparations. Food Control 2022, 135, 108609. [Google Scholar] [CrossRef]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [Green Version]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals; CLSI VET08-ED4:2018; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2018. [Google Scholar]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. 2021. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 3 September 2021).





| Microbial Group | Origin of the Minced Chicken Samples | ||
|---|---|---|---|
| Spain (n = 10) | Portugal (n = 10) | All the Samples (n = 20) | |
| Viable aerobic microbiota | 7.81 ± 0.85 aa | 7.29 ± 1.12 aa | 7.53 ± 1.02 a |
| Psychrotrophic microorganisms | 7.56 ± 0.86 aa | 6.64 ± 1.10 ba | 7.13 ± 1.07 a |
| Enterobacteria | 4.55 ± 0.96 ab | 4.02 ± 0.78 ab | 4.23 ± 0.88 b |
| SAMPLE | Results with q-PCR 1 | Detection with OCLA–PCR 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total Cells | Viable Cells | ||||||||
| Ct 3 | DNA (ng) in the Reaction Tube | Log10 cfu/g in the Sample | Ct PMA | DNA (ng) in the Reaction Tube | Log10 cfu/g in the Sample | % of Total Cells | |||
| SAMPLES FROM SPAIN | CP1 | 38.28 | 0.000012 | 2.72 | 40.00 | 0.000003 | 2.15 | 27.3 | + |
| CP2 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CP3 | 35.26 | 0.000112 | 3.71 | 39.15 | 0.000006 | 2.43 | 5.3 | + | |
| CP4 | 39.72 | 0.000004 | 2.25 | 39.92 | 0.000003 | 2.18 | 86.0 | − | |
| CP5 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CP6 | 33.40 | 0.000458 | 4.32 | 38.37 | 0.000011 | 2.69 | 2.4 | + | |
| CP7 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CP8 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CP9 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CP10 | 36.29 | 0.000052 | 3.37 | 37.59 | 0.000019 | 2.94 | 37.5 | + | |
| SAMPLES FROM PORTUGAL | CPT1 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | − |
| CPT2 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CPT3 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | − | |
| CPT4 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | + | |
| CPT5 | 39.53 | 0.000004 | 2.31 | 40.00 | 0.000003 | 2.15 | 70.2 | + | |
| CPT6 | >40 | <0.000003 | <2.15 | >40 | <0.000003 | <2.15 | - | − | |
| CPT7 | 37.34 | 0.000023 | 3.03 | 40.00 | 0.000003 | 2.15 | 13.5 | + | |
| CPT8 | 33.93 | 0.000307 | 4.14 | 36.67 | 0.000039 | 3.25 | 12.7 | − | |
| CPT9 | 37.33 | 0.000024 | 3.03 | 40.00 | 0.000003 | 2.15 | 13.3 | − | |
| CPT10 | 39.66 | 0.000004 | 2.27 | 40.00 | 0.000003 | 2.15 | 77.4 | + | |
| Antibiotic Resistance Pattern | Number of Isolates | ||
|---|---|---|---|
| From Spain | From Portugal | Total | |
| OX-FOX-FEP | 0 | 1 | 1 |
| OX-FOX-CTX-FEP | 1 | 7 | 8 |
| OX-FOX-CTX-FEP-SXT | 0 | 1 | 1 |
| OX-FOX-CTX-FEP-CIP | 7 | 6 | 13 |
| OX-FOX-CTX-FEP-F | 3 | 3 | 6 |
| OX-FOX-CTX-FEP-E | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-RD | 1 | 1 | 2 |
| OX-FOX-CTX-FEP-SXT-CIP | 2 | 0 | 2 |
| OX-FOX-CTX-FEP-RD-CIP | 3 | 0 | 3 |
| OX-FOX-CTX-FEP-CIP-F | 10 | 4 | 14 |
| OX-FOX-CTX-FEP-E-RD | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-SXT-CIP-F | 2 | 0 | 2 |
| OX-FOX-CTX-FEP-RD-CIP-F | 5 | 1 | 6 |
| OX-FOX-CTX-FEP-E-CIP-F | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-E-RD-CIP | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-SXT-TE-CIP-F | 0 | 1 | 1 |
| OX-FOX-CTX-FEP-CN-E-SXT-CIP | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-E-RD-CIP-F | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-SXT-CIP-ENR-F | 1 | 0 | 1 |
| OX-FOX-CTX-FEP-SXT-RD-CIP-F | 3 | 0 | 3 |
| OX-FOX-CTX-FEP-E-SXT-RD-TE | 1 | 0 | 1 |
| Microbial Group | Culture Media | Incubation | Reference | |
|---|---|---|---|---|
| Time | Temperature (°C) | |||
| Viable aerobic microbiota | PCA 1 | 72 h | 30 °C | [35] |
| Psychrotrophic microorganisms | PCA 1 | 10 days | 7 °C | [84] |
| Enterobacteria | VRBGA 2,3 | 24 h | 37 °C | [85] |
| Gene | Primer | Sequence (5′ → 3′) | Temperature (°C) | Size (bp) |
|---|---|---|---|---|
| lmo1030 | Lmo1030-F | GCTTGTATTCACTTGGATTTGTCTGG | 62 | 509 |
| Lmo1030-R | ACCATCCGCATATCTCAGCCAACT |
| Gene Target | Primer Sequence (5′ → 3′) | Product Size (bp) | Serovar Specificity |
|---|---|---|---|
| lmo0737 | F: AGGGCTTCAAGGACTTACCC R: ACGATTTCTGCTTGCCATTC | 691 | L. monocytogenes serovars 1/2a, 1/2c, 3a, and 3c |
| lmo1118 | F: AGGGGTCTTAAATCCTGGAA R: CGGCTTGTTCGGCATACTTA | 906 | L. monocytogenes serovars 1/2c and 3c |
| ORF2819 | F: AGCAAAATGCCAAAACTCGT R: CATCACTAAAGCCTCCCATTG | 471 | L. monocytogenes serovars 1/2b, 3b, 4b, 4d, and 4e |
| ORF2110 | F: AGTGGACAATTGATTGGTGAA R: CATCCATCCCTTACTTTGGAC | 597 | L. monocytogenes serovars 4b, 4d, and 4e |
| prs | F: GCTGAAGAGATTGCGAAAGAAG R: CAAAGAAACCTTGGATTTGCGG | 370 | All Listeria species |
| Multiplex PCR Fragment Amplification | Serogroup | Listeria monocytogenes Serovar | Control Strain | ||||
|---|---|---|---|---|---|---|---|
| lmo1118 (906 bp) | lmo0737 (691 bp) | ORF2110 (597 bp) | ORF2819 (471 bp) | prs (370 bp) | |||
| − | + | − | − | + | IIa | 1/2a, 3a | ATCC 1 19111 (serovar 1/2a) |
| − | − | − | + | + | IIb | 1/2b, 3b, 7 | STCC 2 936 (serovar 1/2b) |
| + | + | − | − | + | IIc | 1/2c, 3c | STCC 938 (serovar 3c) |
| − | − | − | − | + | IVa | 4a, 4c | ATCC 19114 (serovar 4a) |
| − | − | + | + | + | IVb | 4b, 4d, 4e | ATCC 13932 (serovar 4b) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Melcón, C.; Esteves, A.; Panera-Martínez, S.; Capita, R.; Alonso-Calleja, C. Quantification of Total and Viable Cells and Determination of Serogroups and Antibiotic Resistance Patterns of Listeria monocytogenes in Chicken Meat from the North-Western Iberian Peninsula. Antibiotics 2022, 11, 1828. https://doi.org/10.3390/antibiotics11121828
Rodríguez-Melcón C, Esteves A, Panera-Martínez S, Capita R, Alonso-Calleja C. Quantification of Total and Viable Cells and Determination of Serogroups and Antibiotic Resistance Patterns of Listeria monocytogenes in Chicken Meat from the North-Western Iberian Peninsula. Antibiotics. 2022; 11(12):1828. https://doi.org/10.3390/antibiotics11121828
Chicago/Turabian StyleRodríguez-Melcón, Cristina, Alexandra Esteves, Sarah Panera-Martínez, Rosa Capita, and Carlos Alonso-Calleja. 2022. "Quantification of Total and Viable Cells and Determination of Serogroups and Antibiotic Resistance Patterns of Listeria monocytogenes in Chicken Meat from the North-Western Iberian Peninsula" Antibiotics 11, no. 12: 1828. https://doi.org/10.3390/antibiotics11121828
APA StyleRodríguez-Melcón, C., Esteves, A., Panera-Martínez, S., Capita, R., & Alonso-Calleja, C. (2022). Quantification of Total and Viable Cells and Determination of Serogroups and Antibiotic Resistance Patterns of Listeria monocytogenes in Chicken Meat from the North-Western Iberian Peninsula. Antibiotics, 11(12), 1828. https://doi.org/10.3390/antibiotics11121828