Supramolecular Structure and Renaturation of a (1→3)-β-d-Glucan Compared with Curdlan and Scleroglucan
Abstract
:1. Introduction
2. Experimental Section
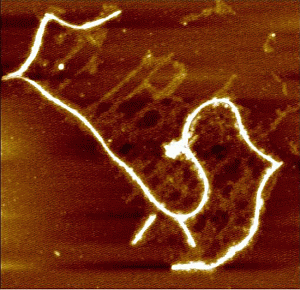
3. Results and Discussion




4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Laroche, C.; Michaud, P. New developments and prospective applications for β-(1,3) glucans. Recent Pat. Biotechnol. 2007, 1, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Survase, S.A.; Saudagar, P.S.; Bajaj, I.B.; Singhal, R.S. Scleroglucan: Fermentative production, downstream processing and applications. Food Technol. Biotecnol. 2007, 45, 107–118. [Google Scholar]
- Duboc, P.; Mollet, B. Applications of exopolysaccharides. Int. Dairy J. 2001, 11, 759–768. [Google Scholar] [CrossRef]
- Sletmoen, M.; Stokke, B.T. Higher order structure of (1,3)-β-d-glucans and its influence on their biological activities and complexation abilities. Biopolymers 2008, 89, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Gawronski, M.; Park, J.T.; Magee, A.S.; Conrad, H. Microfibrillar structure of PGG-glucan in aqueous solution as Triple-helix aggregates by small angle x-ray scattering. Biopolymers 1999, 50, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Masada, M.; Fujimori, K.; Maeda, I. Production of a firm, resilient gel-forming polysaccharide by a mutant of Alcaligenes faecalis var. myxogenes 10C3. Agric. Biol. Chem. 1966, 30, 196–198. [Google Scholar] [CrossRef]
- McIntosh, M.; Stone, B.A.; Stanisich, V.A. Curdlan and other bacterial (1→3)-β-d-glucans. Appl. Microbiol. Biotechnol. 2005, 68, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhang, H.; Yin, Y.; Nishinari, K. Conformation of curdlan as observed by tapping mode atomic force microscopy. Colloid Polym. Sci. 2006, 284, 1371–1377. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Li, Y.; Lv, M.; Li, P.; Xu, H.; Wang, L. Preparation and characterization of novel curdlan/chitosan blending menbranes for antibacterial applications. Carbohydr. Polym. 2011, 84, 952–959. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, L.; Nishinari, K.; Watase, M.; Konno, A. Thermal measurements of curdlan in aqueous suspension during gelation. Food Hydrocolloids 2000, 14, 121–124. [Google Scholar] [CrossRef]
- Vincendon, M. Scleroglucan derivatives: Aromatic carbamates. J. Polym. Sci. Part A Polym. Chem. 1999, 37, 3187–3192. [Google Scholar] [CrossRef]
- Kim, H.I.; Kim, J.H.; Yang, S.B.; Hong, S.G.; Lee, S.A.; Hwang, S.J.; Shin, K.S.; Suh, H.J.; Park, M.H. A polysaccharide extracted from rice bran fermented with Lentinus edodes enhaces natural killer cell activity and exhibits anticancer effects. J. Med. Food 2007, 10, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Palencia, P.; Werning, M.L.; Sierra-Filardi, E.; Dueñas, M.T.; Irastorza, A.; Corbí, A.L.; López, P. Probiotic properties of the 2-substituted (1,3)-β-D-glucan-producing bacterium Pediococcus parvulus 2.6. Appl. Environ. Microbiol. 2009, 75, 4887–4891. [Google Scholar] [CrossRef]
- Mårtensson, O.; Biörklund, M.; Lambo, A.M.; Dueñas-Chasco, M.T.; Irastorza, A.; Holst, O.; Norin, E.; Walling, G.; Öste, R.; Önning, G. Fermented ropy, oat-based products reduce cholesterol levels and stimulate the bifidobacteria flora in humans. Nutr. Res. 2005, 25, 429–442. [Google Scholar]
- Garai-Ibabe, G.; Dueñas, M.T.; Irastorza, A.; Sierra-Filardi, E.; Werning, M.L.; López, P.; Corbí, A.L.; Fernández de Palencia, P. Naturally occurring 2-substituted (1,3)-β-d-glucan producing Lactobacillus suebicus and Pediococcus parvulus strains with potential utility in the production of functional foods. Bioresour. Technol. 2010, 101, 9254–9263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarburu, I. Exopolysaccharide-producing lactic acid bacteria: Structural analysis of polysaccharides and molecular detection of β-(1→3)(1→2)-D-glucan producer strains. Ph.D. Thesis, University of the Basque Country (UPV/EHU), Donostia-San Sebastián, Spain, 2009. [Google Scholar]
- Velasco, S.; Irastorza, A.; Dueñas, M.; Santamaría, A.; Muñoz, M.E. Chemical and rheological properties of the β-glucan produced by Pediococcus parvulus 2.6. J. Agric. Food Chem. 2009, 57, 1827–1834. [Google Scholar] [CrossRef]
- Guenet, J.M. Thermorevesible Gelation of Polymers and Biopolymers; Academic Press Inc.: San Diego, CA, USA, 1992. [Google Scholar]
- Grassi, M.; Lapasin, R.; Pricl, S. A study of the rheological behavior of scleroglucan weak gel systems. Carbohydr. Polym. 1995, 29, 169–181. [Google Scholar] [CrossRef]
- Marieta, C.; Ibarburu, I.; Dueñas, M.; Irastorza, A. Supramolecular structure and conformation of a from a ropy strain of Lactobacillus suebicus as observed by tapping mode atomic force microscopy. J. Agric. Food Chem. 2009, 57, 6183–6188. [Google Scholar] [CrossRef] [PubMed]
- Yanaki, T.; Norisuye, T. Triple helix and random coil scleroglucan in dilute solution. Polym. J. 1983, 15, 389–396. [Google Scholar] [CrossRef]
- Harada, T.; Misaki, A.; Saito, H. Curdlan: A bacterial-gel-forming β-1→3 glucan. Arch. Biochem. Biophys. 1968, 124, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Harada, A. Gel Formation and Ultrastructure in Food Polysaccharides. In Polysaccharide Association Structures in Food; CRC Press: Boca Raton, FL, USA, 1998; pp. 37–40. [Google Scholar]
- Pelosi, P.; Bulone, V.; Heux, L. Polymorphism of curdlan and (1→3)-β-d-glucans synthesized in vitro: A13C CP-MAS and X-Ray diffraction analysis. Carbohydr. Polym. 2006, 66, 199–207. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, C.; Cui, S.; Wang, Z.; Zhang, X. Single–molecules force spectroscopy on curdlan: Unwinding helical structures and random coils. Nanoletters 2003, 3, 1119–1124. [Google Scholar] [CrossRef]
- Nishinari, K. Rheological and DSC study of sol-gel transition in aqueous dispersions of industrially important polymers and colloids. Colloid Polym. Sci. 1997, 275, 1093–1107. [Google Scholar] [CrossRef]
- McIntire, T.M.; Brant, D.A. Imaging of individual biopolymers and supramolecular assemblies using noncontact atomic force microscopy. Biopolymers 1997, 42, 133–146. [Google Scholar] [CrossRef] [PubMed]
- McIntire, T.M.; Penner, R.M.; Brant, D.A. Observations of a circular, triple-helical polysaccharide using noncontact atomic force microscopy. Macromolecules 1995, 28, 6375–6377. [Google Scholar] [CrossRef]
- Vuppu, A.; Garcia, A.A.; Vernia, C. Tapping mode atomic force microscopy of scleroglucan networks. Biopolymers 1997, 42, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Stokke, B.T.; Elgsaeter, A.; Brant, D.A.; Kuge, T.; Kitamura, S. Macromolecular cyclization of (1→6)-branched-(1→3)-β-d-glucans observed after denaturation-renaturation of the triple-helical structure. Biopolymers 1993, 33, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Brant, D.A. Versatile polysaccharides: Ropes, rings, and rods. AIM Mag. 2005, 60, 40–49. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Puertas, A.I.; Dueñas, M.T.; Marieta, C. Supramolecular Structure and Renaturation of a (1→3)-β-d-Glucan Compared with Curdlan and Scleroglucan. Fibers 2014, 2, 255-263. https://doi.org/10.3390/fib2030255
Puertas AI, Dueñas MT, Marieta C. Supramolecular Structure and Renaturation of a (1→3)-β-d-Glucan Compared with Curdlan and Scleroglucan. Fibers. 2014; 2(3):255-263. https://doi.org/10.3390/fib2030255
Chicago/Turabian StylePuertas, Ana Isabel, Mª Teresa Dueñas, and Cristina Marieta. 2014. "Supramolecular Structure and Renaturation of a (1→3)-β-d-Glucan Compared with Curdlan and Scleroglucan" Fibers 2, no. 3: 255-263. https://doi.org/10.3390/fib2030255
APA StylePuertas, A. I., Dueñas, M. T., & Marieta, C. (2014). Supramolecular Structure and Renaturation of a (1→3)-β-d-Glucan Compared with Curdlan and Scleroglucan. Fibers, 2(3), 255-263. https://doi.org/10.3390/fib2030255