Zinc-Dependent Oligomerization of Thermus thermophilus Trigger Factor Chaperone
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Protein Expression and Purification
2.3. Preparation of Zinc-Bound and Zinc-Depleted Proteins
2.4. MALDI-TOF-MS Spectrometry
2.5. Circular Dichroism Measurement
2.6. Size Exclusion Chromatography in a Multi-Angle Laser Light Scattering (SEC-MALS)
2.7. NMR Spectroscopy
3. Results
3.1. Zinc-Binding Is Characteristic to TtTF
3.2. Full-Length of TtTF Was Required for Zinc Recognition
3.3. Zn2+ Induced Little Effect to Thermal Stability of TtTF
3.4. Zn2+ Induced Partial Structural Change of TtTF
3.5. Zn2+ Promoted Oligomerization of TtTF
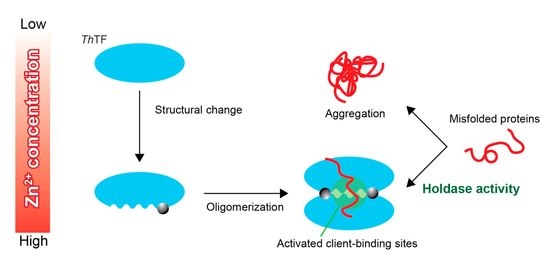
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobson, C.M. Protein Folding and Misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Rabbani, G.; Choi, I. Roles of Osmolytes in Protein Folding and Aggregation in Cells and Their Biotechnological Applications. Int. J. Biol. Macromol. 2018, 109, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular Chaperones and Protein Quality Control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Hartl, F.U.; Hayer-Hartl, M. Molecular Chaperones in the Cytosol: From Nascent Chain to Folded Protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saibil, H. Chaperone Machines for Protein Folding, Unfolding and Disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Agashe, V.R.; Guha, S.; Chang, H.; Genevaux, P.; Hayer-hartl, M.; Stemp, M.; Georgopoulos, C.; Hartl, F.U.; Barral, J.M. Function of Trigger Factor and DnaK in Multidomain Protein Folding: Increase in Yield at the Expense of Folding Speed. Cell 2004, 117, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.; Bukau, B.; Kramer, G. Structure and Function of the Molecular Chaperone Trigger Factor. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Crooke, E.; Wickner, W. Trigger Factor: A Soluble Protein That Folds pro-OmpA into a Membrane-Assembly-Competent Form. Proc. Natl. Acad. Sci. USA 1987, 84, 5216–5220. [Google Scholar] [CrossRef] [Green Version]
- De Geyter, J.; Portaliou, A.G.; Srinivasu, B.; Krishnamurthy, S.; Economou, A.; Karamanou, S. Trigger Factor Is a Bona Fide Secretory Pathway Chaperone That Interacts with SecB and the Translocase. EMBO Rep. 2020, 21, 1–17. [Google Scholar] [CrossRef]
- Kandror, O.; Sherman, M.; Rhode, M.; Goldberg, A.L. Trigger Factor Is Involved in GroEL-Dependent Protein Degradation in Escherichia coli and Promotes Binding of GroEL to Unfolded Proteins. EMBO J. 1995, 14, 6021–6027. [Google Scholar] [CrossRef] [PubMed]
- Rizzolo, K.; Yu, A.Y.H.; Ologbenla, A.; Kim, S.R.; Zhu, H.; Ishimori, K.; Thibault, G.; Leung, E.; Zhang, Y.W.; Teng, M.; et al. Functional Cooperativity between the Trigger Factor Chaperone and the ClpXP Proteolytic Complex. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Ferbitz, L.; Maier, T.; Patzelt, H.; Bukau, B.; Deuerling, E.; Ban, N. Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins. Nature 2004, 431, 590–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saio, T.; Guan, X.; Rossi, P.; Economou, A.; Kalodimos, C.G. Structural Basis for Protein Antiaggregation Activity of the Trigger Factor Chaperone. Science 2014, 344, 590–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, A.; Becker, A.H.; Zachmann-Brand, B.; Deuerling, E.; Bukau, B.; Kramer, G. Concerted Action of the Ribosome and the Associated Chaperone Trigger Factor Confines Nascent Polypeptide Folding. Mol. Cell 2012, 48, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Maier, R.; Eckert, B.; Scholz, C.; Lilie, H.; Schmid, F.X. Interaction of Trigger Factor with the Ribosome. J. Mol. Biol. 2003, 326, 585–592. [Google Scholar] [CrossRef]
- Huang, C.; Rossi, P.; Saio, T.; Kalodimos, C.G. Structural Basis for the Antifolding Activity of a Molecular Chaperone. Nature 2016, 537, 202–206. [Google Scholar] [CrossRef] [Green Version]
- Saio, T.; Kawagoe, S.; Ishimori, K.; Kalodimos, C.G. Oligomerization of a Molecular Chaperone Modulates Its Activity. Elife 2018, 7, 1–18. [Google Scholar] [CrossRef]
- Morgado, L.; Burmann, B.M.; Sharpe, T.; Mazur, A.; Hiller, S. The Dynamic Dimer Structure of the Chaperone Trigger Factor. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linke, K.; Wolfram, T.; Bussemer, J.; Jakob, U. The Roles of the Two Zinc Binding Sites in DnaJ. J. Biol. Chem. 2003, 278, 44457–44466. [Google Scholar] [CrossRef] [Green Version]
- Suno, R.; Taguchi, H.; Masui, R.; Odaka, M.; Yoshida, M. Trigger Factor from Thermus Thermophilus Is a Zn2+-Dependent Chaperone. J. Biol. Chem. 2004, 279, 6380–6384. [Google Scholar] [CrossRef] [Green Version]
- Kawagoe, S.; Nakagawa, H.; Kumeta, H.; Ishimori, K.; Saio, T. Structural Insight into Proline Cis/Trans Isomerization of Unfolded Proteins Catalyzed by the Trigger Factor Chaperone. J. Biol. Chem. 2018, 293, 15095–15106. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.A.; Chacón, P.; Merelo, J.J.; Morán, F. Evaluation of Secondary Structure of Proteins from UV Circular Dichroism Spectra Using an Unsupervised Learning Neural Network. Protein Eng. Des. Sel. 1993, 6, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a Website for Calculating Protein Secondary Structure from Circular Dichroism Spectroscopic Data. Protein Sci. 2021, 1–10. [Google Scholar] [CrossRef]
- Miles, A.J.; Whitmore, L.; Wallace, B.A. Spectral Magnitude Effects on the Analyses of Secondary Structure from Circular Dichroism Spectroscopic Data. Protein Sci. 2005, 14, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Ullah, Z.; Iqbal, A.; Baloch, M.K.; Nishan, U.; Shah, M. New Insights into the Zinc- α2-Glycoprotein (ZAG) Scaffold and Its Metal Ions Binding Abilities Using Spectroscopic Techniques. Life Sci. 2020, 249, 117462. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ariza, J.L.; García-Barrera, T.; Lorenzo, F.; Bernal, V.; Villegas, M.J.; Oliveira, V. Use of Mass Spectrometry Techniques for the Characterization of Metal Bound to Proteins (Metallomics) in Biological Systems. Anal. Chim. Acta 2004, 524, 15–22. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Hirel, P.; Schmitter, J.M.; Dessen, P.; Fayat, G.; Blanquet, S. Extent of N-terminal Methionine Excision from Escherichia coli Proteins Is Governed by the Side-chain Length of the Penultimate Amino Acid. Proc. Natl. Acad. Sci. USA 1989, 86, 8247–8251. [Google Scholar] [CrossRef] [Green Version]
- Oshima, T.; Imahori, K. Description of Thermus Thermophilus (Yoshida and Oshima) Comb. Nov., a Nonsporulating Thermophilic Bacterium from a Japanese Thermal Spa. Int. J. Syst. Bacteriol. 1974, 24, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Benjwal, S.; Verma, S.; Röhm, K.H.; Gursky, O. Monitoring Protein Aggregation during Thermal Unfolding in Circular Dichroism Experiments. Protein Sci. 2006, 15, 635–639. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Collected as a Function of Temperature to Determine the Thermodynamics of Protein Unfolding and Binding Interactions. Nat. Protoc. 2007, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Patzelt, H.; Kramer, G.; Rauch, T.; Schönfeld, H.J.; Bukau, B.; Deuerling, E. Three-State Equilibrium of Escherichia coli Trigger Factor. Biol. Chem. 2002, 383, 1611–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludlam, A.V.; Moore, B.A.; Xu, Z. The Crystal Structure of Ribosomal Chaperone Trigger Factor from Vibrio Cholerae. Proc. Natl. Acad. Sci. USA 2004, 101, 13436–13441. [Google Scholar] [CrossRef] [Green Version]
- Fujino, Y.; Miyagawa, T.; Torii, M.; Inoue, M.; Fujii, Y.; Okanishi, H.; Kanai, Y.; Masui, R. Structural Changes Induced by Ligand Binding Drastically Increase the Thermostability of the Ser/Thr Protein Kinase TpkD from Thermus Thermophilus HB8. FEBS Lett. 2021, 595, 264–274. [Google Scholar] [CrossRef]
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of Metal in Folding and Stability of Copper Proteins in Vitro. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 1594–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojoh, K.; Matsuzawa, H.; Wakagi, T. Zinc and an N-Terminal Extra Stretch of the Ferredoxin from a Thermoacidophilic Archaeon Stabilize the Molecule at High Temperature. Eur. J. Biochem. 1999, 264, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Ave ± SD (Da) (Refolded in Zn2+ Containing Buffer) | Ave ± SD (Da) (Refolded in EDTA Containing Buffer) | δ (Da) | |
|---|---|---|---|
| TtTF | 46381 ± 8 | 46319 ± 4 | 62 ± 12 |
| TmTF | 51338 ± 13 | 51335 ± 12 | 3 ± 25 |
| EcTF | 48218 ± 12 | 48214 ± 11 | 4 ± 22 |
| TtTFRBD | 12698 ± 2 | 12702 ± 3 | −4 ± 5 |
| TtTFSBD | 25646 ± 5 | 25648 ± 2 | −2 ± 6 |
| TtTFPPD | 8899 ± 3 | 8899 ± 2 | 0 ± 4 |
| TtTFPPD-SBD | 35990 ± 6 | 36005 ± 4 | −15 ± 10 |
| TtTFRBD-SBD | 40075 ± 4 | 40098 ± 6 | −23 ± 9 |
| Temperature (°C) | State of TtTF | α-Helix | β-Sheet | Random Coil |
|---|---|---|---|---|
| 20 | TtTF (EDTA) | 56% | 9% | 35% |
| TtTF (Zn2+) | - | - | - | |
| 35 | TtTF (EDTA) | 59% | 8% | 34% |
| TtTF (Zn2+) | 47% | 18% | 35% | |
| 50 | TtTF (EDTA) | 54% | 10% | 36% |
| TtTF (Zn2+) | 46% | 17% | 37% | |
| 65 | TtTF (EDTA) | - | - | - |
| TtTF (Zn2+) | 42% | 20% | 37% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Matsusaki, M.; Sugawara, T.; Ishimori, K.; Saio, T. Zinc-Dependent Oligomerization of Thermus thermophilus Trigger Factor Chaperone. Biology 2021, 10, 1106. https://doi.org/10.3390/biology10111106
Zhu H, Matsusaki M, Sugawara T, Ishimori K, Saio T. Zinc-Dependent Oligomerization of Thermus thermophilus Trigger Factor Chaperone. Biology. 2021; 10(11):1106. https://doi.org/10.3390/biology10111106
Chicago/Turabian StyleZhu, Haojie, Motonori Matsusaki, Taiga Sugawara, Koichiro Ishimori, and Tomohide Saio. 2021. "Zinc-Dependent Oligomerization of Thermus thermophilus Trigger Factor Chaperone" Biology 10, no. 11: 1106. https://doi.org/10.3390/biology10111106
APA StyleZhu, H., Matsusaki, M., Sugawara, T., Ishimori, K., & Saio, T. (2021). Zinc-Dependent Oligomerization of Thermus thermophilus Trigger Factor Chaperone. Biology, 10(11), 1106. https://doi.org/10.3390/biology10111106