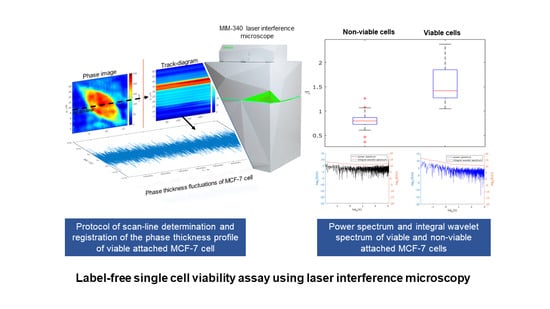
Label-Free Single Cell Viability Assay Using Laser Interference Microscopy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Culture Conditions
2.2. Cell Sample Preparation
2.2.1. Suspension of Viable Cells
2.2.2. Suspension of Non-Viable Cells
2.2.3. Viable Cells Attached to a Glass Surface
2.2.4. Non-Viable Cells Attached to a Glass Surface
2.3. Trypan Blue Staining
2.4. Cell Research by LIM
2.4.1. Technical Characteristics of the Laser Interference Microscope MIM-340
2.4.2. MIM-340 Laser Measurement of Phase Thickness Fluctuations
2.4.3. Phase Thickness Fluctuations Analysis
2.4.4. Statistical Analysis
3. Results and Discussion
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, M.; Sun, J.; Tang, Y.; Dong, B.; Lin, W. Discriminating Live and Dead Cells in Dual-Color Mode with a Two-Photon Fluorescent Probe Based on ESIPT Mechanism. Anal. Chem. 2018, 90, 998–1005. [Google Scholar] [CrossRef]
- Stoddart, M.J. Cell Viability Assays: Introduction BT–Mammalian Cell Viability: Methods and Protocols. In Mammalian Cell Viability; Humana Press: Totowa, NJ, USA, 2011; pp. 1–6. [Google Scholar]
- Fotakis, G.; Timbrell, J.A. In Vitro Cytotoxicity Assays: Comparison of LDH, Neutral Red, MTT and Protein Assay in Hepatoma Cell Lines Following Exposure to Cadmium Chloride. Toxicol. Lett. 2006, 160, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, D.H.; Abrahamse, H. The Role of Laser Fluence in Cell Viability, Proliferation, and Membrane Integrity of Wounded Human Skin Fibroblasts Following Helium-Neon Laser Irradiation. Lasers Surg. Med. 2006, 38, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Van Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cancer Cell Culture: Cell Sensitivity Assays. MTT Assay 2011, 731, 237–345. [Google Scholar] [CrossRef]
- An, W.F.; Tolliday, N.J. Introduction: Cell-Based Assays for High-Throughput Screening; Humana Press: Totowa, NJ, USA, 2009; pp. 1–12. [Google Scholar] [CrossRef]
- Atale, N.; Gupta, S.; Yadav, U.C.S.; Rani, V. Cell-Death Assessment by Fluorescent and Nonfluorescent Cytosolic and Nuclear Staining Techniques. J. Microsc. 2014, 255, 7–19. [Google Scholar] [CrossRef]
- Johnson, S.; Nguyen, V.; Coder, D. Assessment of Cell Viability. Curr. Protoc. Cytom. 2013, 1, 9.2.1–9.2.26. [Google Scholar] [CrossRef] [PubMed]
- Kaluzki, I.; Hailemariam-Jahn, T.; Doll, M.; Kaufmann, R.; Balermpas, P.; Zöller, N.; Kippenberger, S.; Meissner, M. Dimethylfumarate Inhibits Colorectal Carcinoma Cell Proliferation: Evidence for Cell Cycle Arrest, Apoptosis and Autophagy. Cells 2019, 8, 1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanipandian, N.; Li, D.; Kannan, S. Induction of Intrinsic Apoptotic Signaling Pathway in A549 Lung Cancer Cells Using Silver Nanoparticles from Gossypium Hirsutum and Evaluation of in Vivo Toxicity. Biotechnol. Rep. 2019, 23, e00339. [Google Scholar] [CrossRef] [PubMed]
- Kudriavtsev, I.V.; Golovkin, A.S.; Zurochka, A.V.; Khaidukov, S.V. Modern Technologies and Approaches to Apoptosis Studies in Experimental Biology. Med. Immunol. 2014, 14, 461–482. [Google Scholar] [CrossRef] [Green Version]
- Newton, R.; Delguste, M.; Koehler, M.; Dumitru, A.C.; Laskowski, P.R.; Müller, D.J.; Alsteens, D. Combining Confocal and Atomic Force Microscopy to Quantify Single-Virus Binding to Mammalian Cell Surfaces. Nat. Protoc. 2017, 12, 2275–2292. [Google Scholar] [CrossRef] [PubMed]
- Sandin, J.N.; Aryal, S.P.; Wilkop, T.; Richards, C.I.; Grady, M.E. Near Simultaneous Laser Scanning Confocal and Atomic Force Microscopy (Conpokal) on Live Cells. J. Vis. Exp. 2020, 2020, 1–25. [Google Scholar] [CrossRef]
- Andresen, V.; Pollok, K.; Rinnenthal, J.L.; Oehme, L.; Günther, R.; Spiecker, H.; Radbruch, H.; Gerhard, J.; Sporbert, A.; Cseresnyes, Z.; et al. High-Resolution Intravital Microscopy. PLoS ONE 2012, 7, e50915. [Google Scholar] [CrossRef] [Green Version]
- Coste, A.; Oktay, M.H.; Condeelis, J.S.; Entenberg, D. Intravital Imaging Techniques for Biomedical and Clinical Research. Cytom. Part A 2020, 97, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, 2016, 343–346. [Google Scholar] [CrossRef]
- Simon, A.; Cohen-Bouhacina, T.; Porté, M.C.; Aimé, J.P.; Amédée, J.; Bareille, R.; Baquey, C. Characterization of Dynamic Cellular Adhesion of Osteoblasts Using Atomic Force Microscopy. Cytom. Part A 2003, 54, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L.; Markossian, S.; Sittampalam, G.S.; Grossman, A.; et al. Cell viablity assays. In Assay Guidance Manual; Markosian, S., Sittampalam, G.S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Caaveiro, J.M.M., Chung, T.D.Y., et al., Eds.; Eli Lilly & Company and the National Centre for Advancing Translational Sciences: Bethesda, MD, USA, 2013; Updated 2016. [Google Scholar]
- Aslantürk, Ö.S. In Vitro Cytotoxicity and Cell Viability Assays: Principles, Advantages, and Disadvantages. In Genotoxicity A Predictable Risk to Our Actual World; InTech: Rijeka, Croatia, 2018. [Google Scholar] [CrossRef] [Green Version]
- Wojcik, K.; Dobrucki, J.W. Interaction of a DNA Intercalator DRAQ5, and a Minor Groove Binder SYTO17, with Chromatin in Live Cells–Influence on Chromatin Organization and Histone–DNA Interactions. Cytom. Part A 2008, 73, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P. Photobleaching of Organic Fluorophores: Quantitative Characterization, Mechanisms, Protection. Methods Appl. Fluoresc. 2020, 8, 22001. [Google Scholar] [CrossRef]
- Frigault, M.M.; Lacoste, J.; Swift, J.L.; Brown, C.M. Live-Cell Microscopy–Tips and Tools. J. Cell Sci. 2009, 122, 753–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Song, Z.; Liu, Q.; Song, Y. Recent advances in dielectrophoresis-based cell viability assessment. Electrophoresis 2020, 41, 917–932. [Google Scholar] [CrossRef] [PubMed]
- Duellman, S.J.; Zhou, W.; Meisenheimer, P.; Vidugiris, G.; Cali, J.J.; Gautam, P.; Wennerberg, K.; Vidugiriene, J. Bioluminescent, Nonlytic, Real-Time Cell Viability Assay and Use in Inhibitor Screening. Assay Drug Dev. Technol. 2015, 13, 456–465. [Google Scholar] [CrossRef] [Green Version]
- Yusipovich, A.I.; Parshina, E.Y.; Brysgalova, N.Y.; Brazhe, A.R.; Brazhe, N.A.; Lomakin, A.G.; Levin, G.G.; Maksimov, G.V. Laser Interference Microscopy in Erythrocyte Study. J. Appl. Phys. 2009, 105. [Google Scholar] [CrossRef]
- Majeed, H.; Sridharan, S.; Mir, M.; Ma, L.; Min, E.; Jung, W.; Popescu, G. Quantitative Phase Imaging for Medical Diagnosis. J. Biophotonics 2017, 10, 177–205. [Google Scholar] [CrossRef]
- Ignatyev, P.S.; Indukaev, K.V.; Osipov, P.A.; Sergeev, I.K. Laser Interference Microscopy for Nanobiotechnologies. Biomed. Eng. 2013, 47, 32–35. [Google Scholar] [CrossRef]
- Tychinskii, V.P. Dynamic Phase Microscopy: Is a “Dialogue” with the Cell Possible? Phys. Usp. 2007, 50, 513–528. [Google Scholar] [CrossRef]
- Loparev, A.V.; Ignat’Ev, P.S.; Indukaev, K.V.; Osipov, P.A.; Mazalov, I.N.; Kozyrev, A.V. A High-Speed Modulation Interference Microscope for Biomedical Studies. Meas. Tech. 2009, 52, 1229–1235. [Google Scholar] [CrossRef]
- Kaplonek, W.; Lukianowicz, C. Coherence Correlation Interferometry in Surface Topography Measurements. In Recent Interferometry Applications in Topography and Astronomy; Padron, I., Ed.; InTech: Rijeka, Croatia, 2012; pp. 1–26. [Google Scholar] [CrossRef] [Green Version]
- Nebogatikov, V.; Nikitiuk, A.; Konysheva, A.; Ignatyev, P.; Grishko, V.; Naimark, O. Study of Morphological Changes in Breast Cancer Cells MCF-7 under the Action of pro-Apoptotic Agents with Laser Modulation Interference Microscope MIM-340. AIP Conf. Proc. 2017, 1882, 20053. [Google Scholar] [CrossRef]
- Nikitiuk, A.S.; Voronina, A.O.; Beloglazova, Y.A.; Ozernykh, O.S.; Grishko, V.V.; Naimark, O.B. Study of Cancer and Normal Cells Dynamics Based on Laser Interference Microscopy. AIP Conf. Proc. 2020, 2216, 60005. [Google Scholar] [CrossRef]
- Naimark, O.B. Structural-Scaling Transitions and Localized Distortion Modes in the DNA Double Helix. Phys. Mesomech. 2007, 10, 33–45. [Google Scholar] [CrossRef]
- Bizzarri, M.; Naimark, O.; Nieto-Villar, J.; Fedeli, V.; Giuliani, A. Complexity in Biological Organization: Deconstruction (and Subsequent Restating) of Key Concepts. Entropy 2020, 22, 885–900. [Google Scholar] [CrossRef] [PubMed]
- Naimark, O. Mesoscopic Cell Dynamics in Different Environment and Problem of Cancer. AIP Conf. Proc. 2019, 2167, 20237. [Google Scholar] [CrossRef]
- Gerasimova, E.; Audit, B.; Roux, S.G.; Khalil, A.; Gileva, O.; Argoul, F.; Naimark, O.; Arneodo, A. Wavelet-Based Multifractal Analysis of Dynamic Infrared Thermograms to Assist in Early Breast Cancer Diagnosis. Front. Physiol. 2014, 5, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soule, H.D.; Vazquez, J.; Long, A.; Albert, S.; Brennan, M. A Human Cell Line from a Pleural Effusion Derived from a Breast carcinoma1,2. J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 Years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the Right Cell Line for Breast Cancer Research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.V.; Oesterreich, S.; Davidson, N.E. MCF-7 Cells–Changing the Course of Breast Cancer Research and Care for 45 Years. J. Natl. Cancer Inst. 2015, 107, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelfand, R.; Vernet, D.; Bruhn, K.W.; Sarkissyan, S.; Heber, D.; Vadgama, J.V.; Gonzalez-Cadavid, N.F. Long-Term Exposure of MCF-7 Breast Cancer Cells to Ethanol Stimulates Oncogenic Features. Int. J. Oncol. 2017, 50, 49–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emily Joo, E.; Yamada, K.M. Cell Adhesion and Movement. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Academic Press: Cambridge, MA, USA, 2015; pp. 61–72. [Google Scholar] [CrossRef]
- Rappaz, B.; Marquet, P.; Cuche, E.; Emery, Y.; Depeursinge, C.; Magistretti, P.J. Measurement of the Integral Refractive Index and Dynamic Cell Morphometry of Living Cells with Digital Holographic Microscopy. Opt. Express 2005, 13, 9361–9373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.R.; Kim, K.; Jung, J.; Heo, J.H.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.J.; Park, H.; Park, Y.K. Quantitative Phase Imaging Techniques for the Study of Cell Pathophysiology: From Principles to Applications. Sensors 2013, 13, 4170–4191. [Google Scholar] [CrossRef]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting Nanoscale Vibrations as Signature of Life. Proc. Natl. Acad. Sci. USA. 2015, 112, 378–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, A.C.; Venturelli, L.; Longo, G.; Dietler, G.; Kasas, S. Nanomotion Detection Based on Atomic Force Microscopy Cantilevers. Cell Surf. 2019, 5, 100021. [Google Scholar] [CrossRef]
- Wu, S.; Liu, X.; Zhou, X.; Liang, X.M.; Gao, D.; Liu, H.; Zhao, G.; Zhang, Q.; Wu, X. Quantification of Cell Viability and Rapid Screening Anti-Cancer Drug Utilizing Nanomechanical Fluctuation. Biosens. Bioelectron. 2016, 77, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.C.; Rosenblum, M.G.; Peng, C.K.; Mietus, J.; Havlin, S.; Stanley, H.E.; Goldberger, A.L. Scaling Behaviour of Heartbeat Intervals Obtained by Wavelet-Based Time- Series Analysis. Nature 1996, 383, 323–327. [Google Scholar] [CrossRef]
- Thurner, S.; Feurstein, M.C.; Teich, M.C. Multiresolution Wavelet Analysis of Heartbeat Intervals Discriminates Healthy Patients from Those with Cardiac Pathology. Phys. Rev. Lett. 1998, 80, 1544–1547. [Google Scholar] [CrossRef] [Green Version]
- Pavlov, A.N.; Ziganshin, A.R.; Klimova, O.A. Multifractal Characterization of Blood Pressure Dynamics: Stress-Induced Phenomena. Chaos Solitons Fractals 2005, 24, 57–63. [Google Scholar] [CrossRef]
- Marrone, A.; Polosa, A.D.; Scioscia, G.; Stramaglia, S.; Zenzola, A. Multiscale Analysis of Blood Pressure Signals. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 1999, 60, 1088–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicar, T.; Raudenska, M.; Gumulec, J.; Balvan, J. The Quantitative-Phase Dynamics of Apoptosis and Lytic Cell Death. Sci. Rep. 2020, 10, 1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ash, W.M.; Krzewina, L.; Kim, M.K. Quantitative Imaging of Cellular Adhesion by Total Internal Reflection Holographic Microscopy. Appl. Opt. 2009, 48, H144–H152. [Google Scholar] [CrossRef]
- Yang, F.; Riedel, R.; Pino, P.; Pelaz, B.; Said, A.H.; Soliman, M.; Pinnapireddy, S.R.; Feliu, N.; Parak, W.J.; Bakowsky, U.; et al. Real-Time, Label-Free Monitoring of Cell Viability Based on Cell Adhesion Measurements with an Atomic Force Microscope. J. Nanobiotechnology 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drumheller, P.D.; Hubbell, J.A. Surface immobilization of adhesion ligands for investigations of cell-substrate interactions. Biomed. Eng. Handb. 2000, 2, 1583–1596. [Google Scholar]
- Naimark, O.B.; Bayandin, Y.V.; Grishko, V.V.; Nikitiuk, A.S. Mesoscopic Cell Mechanobiology and the Problem of Cancer. Org. J. Biol. Sci. 2020, 4, 42–56. [Google Scholar] [CrossRef]
- Bayandin, Y.V.; Nikitiuk, A.S.; Voronina, A.O.; Beloglazova, Y.A.; Gagarskikh, O.N.; Grishko, V.V.; Naimark, O.B. Analysis of Morphometric and Fractal Properties of Phase-Contrast Images of Native and Apoptotically Changed MCF-7 Cells. Russ. J. Biomech. 2020, 24, 268–276. [Google Scholar] [CrossRef]
- Naimark, O.B.; Bayandin, Y.V.; Beloglazova, Y.A.; Gagarskich, O.N.; Grishko, V.V.; Nikitiuk, A.S.; Voronina, A.O. DNA Transformation, Cell Epigenetic Landscape and Open Complex Dynamics in Cancer Development. Math. Biol. Bioinform. 2020, 15, 251–267. [Google Scholar] [CrossRef]







| Percentiles | Attached Viable Cells (Sample Nr.1) | Attached Non-Viable Cells (Sample Nr.2) | Suspended Viable Cells (Sample Nr.3) | Suspended Non-Viable Cells (Sample Nr.4) |
|---|---|---|---|---|
| 25th | 21.2 | 9.9 | 7.0 | 7.6 |
| 75th | 111.6 | 20.6 | 16.0 | 12.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beloglazova, Y.; Nikitiuk, A.; Voronina, A.; Gagarskikh, O.; Bayandin, Y.; Naimark, O.; Grishko, V. Label-Free Single Cell Viability Assay Using Laser Interference Microscopy. Biology 2021, 10, 590. https://doi.org/10.3390/biology10070590
Beloglazova Y, Nikitiuk A, Voronina A, Gagarskikh O, Bayandin Y, Naimark O, Grishko V. Label-Free Single Cell Viability Assay Using Laser Interference Microscopy. Biology. 2021; 10(7):590. https://doi.org/10.3390/biology10070590
Chicago/Turabian StyleBeloglazova, Yulia, Aleksandr Nikitiuk, Anna Voronina, Olga Gagarskikh, Yuriy Bayandin, Oleg Naimark, and Victoria Grishko. 2021. "Label-Free Single Cell Viability Assay Using Laser Interference Microscopy" Biology 10, no. 7: 590. https://doi.org/10.3390/biology10070590
APA StyleBeloglazova, Y., Nikitiuk, A., Voronina, A., Gagarskikh, O., Bayandin, Y., Naimark, O., & Grishko, V. (2021). Label-Free Single Cell Viability Assay Using Laser Interference Microscopy. Biology, 10(7), 590. https://doi.org/10.3390/biology10070590