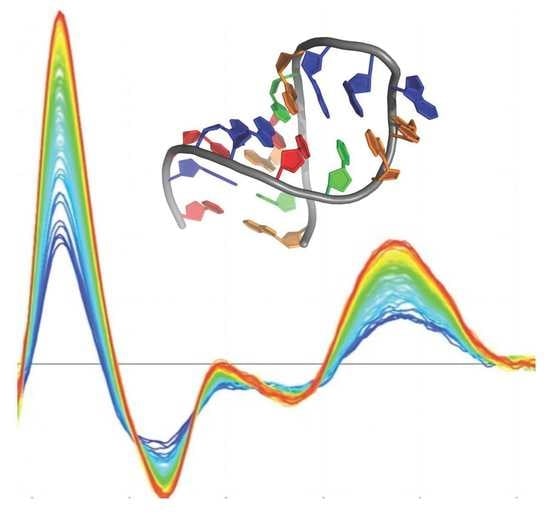
The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Hfq CTR Peptide and Protein
2.3. DsrA and rpoS sequences
2.4. Synchrotron Radiation Circular Dichroism (SRCD)
2.5. ElectroMobility Shift Assay (EMSA)
3. Results
3.1. The Amyloid CTR Region of Hfq Triggers RNA Annealing
3.2. Hfq-CTR Stabilizes DsrA Secondary Structure
3.3. Hfq CTR Does Not Affect rpoS Secondary Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CTR/NTR | C/N-terminal region |
| IDP | Intrinsically Disordered Proteins |
| rbs | ribosome binding site |
| sRNA | small noncoding RNAs |
| SL | stem-loop |
| SRCD | Synchrotron Radiation Circular Dichroism. |
References
- Beales, N. Adaptation of Microorganisms to Cold Temperatures, Weak Acid Preservatives, Low pH, and Osmotic Stress: A Review. Compr. Rev. Food Sci. Food Saf. 2004, 3, 1–20. [Google Scholar] [CrossRef]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [Green Version]
- Gottesman, S.; McCullen, C.A.; Guillier, M.; Vanderpool, C.K.; Majdalani, N.; Benhammou, J.; Thompson, K.M.; FitzGerald, P.C.; Sowa, N.A.; FitzGerald, D.J. Small RNA regulators and the bacterial response to stress. Compr. Rev. Food Sci. Food Saf. 2006, 71, 1–11. [Google Scholar] [CrossRef]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef]
- De Lay, N.; Schu, D.J.; Gottesman, S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol. Chem. 2013, 288, 7996–8003. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Brennan, R.G.; Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007, 10, 125–133. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. RNA reflections: Converging on Hfq. RNA 2015, 21, 511–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobrero, P.; Valverde, C. The bacterial protein Hfq: Much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012, 38, 276–299. [Google Scholar] [CrossRef] [PubMed]
- Franze de Fernandez, M.T.; Eoyang, L.; August, J.T. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 1968, 219, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Franze de Fernandez, M.T.; Hayward, W.S.; August, J.T. Bacterial proteins required for replication of phage Qb ribonucleic acid. J. Biol. Chem. 1972, 247, 824–831. [Google Scholar] [CrossRef]
- Sun, X.; Zhulin, I.; Wartell, R.M. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002, 30, 3662–3671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Muffler, A.; Traulsen, D.D.; Fischer, D.; Lange, R.; Hengge-Aronis, R. The RNA-binding protein HF-1 plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sSsubunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1997, 179, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilusz, C.J.; Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005, 12, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wilusz, J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013, 10, 592–601. [Google Scholar] [CrossRef] [Green Version]
- Moller, T.; Franch, T.; Hojrup, P.; Keene, D.R.; Bachinger, H.P.; Brennan, R.G.; Valentin-Hansen, P. Hfq: A Bacterial Sm-like Protein that Mediates RNA-RNA Interaction. Mol. Cell 2002, 9, 23–30. [Google Scholar] [PubMed]
- Sauter, C.; Basquin, J.; Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003, 31, 4091–4098. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Pearson, R.F.; Moller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: A bacterial Sm-like protein. EMBO J. 2002, 21, 3546–3556. [Google Scholar] [CrossRef] [Green Version]
- Arluison, V.; Folichon, M.; Marco, S.; Derreumaux, P.; Pellegrini, O.; Seguin, J.; Hajnsdorf, E.; Regnier, P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004, 271, 1258–1265. [Google Scholar] [CrossRef]
- Robinson, K.E.; Orans, J.; Kovach, A.R.; Link, T.M.; Brennan, R.G. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014, 42, 2736–2749. [Google Scholar] [CrossRef]
- Sauer, E.; Schmidt, S.; Weichenrieder, O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 9396–9401. [Google Scholar] [CrossRef] [Green Version]
- Updegrove, T.B.; Zhang, A.; Storz, G. Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 2016, 30, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimastrogiovanni, D.; Frohlich, K.S.; Bandyra, K.J.; Bruce, H.A.; Hohensee, S.; Vogel, J.; Luisi, B.F. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife 2014, 3, e05375. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Curtis, J.E.; Fang, X.; Woodson, S.A. Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc. Natl. Acad. Sci. USA 2014, 111, 17134–17139. [Google Scholar] [CrossRef] [Green Version]
- Udekwu, K.I.; Darfeuille, F.; Vogel, J.; Reimegard, J.; Holmqvist, E.; Wagner, E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005, 19, 2355–2366. [Google Scholar] [CrossRef] [Green Version]
- Moon, K.; Gottesman, S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011, 13, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Wassarman, K.M.; Ortega, J.; Steven, A.C.; Storz, G. The Sm-like Hfq Protein Increases OxyS RNA Interaction with Target mRNAs. Mol. Cell 2002, 9, 11–22. [Google Scholar] [CrossRef]
- Aiba, H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007, 10, 134–139. [Google Scholar] [CrossRef]
- Sauer, E.; Weichenrieder, O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. USA 2011, 108, 13065–13070. [Google Scholar] [CrossRef] [Green Version]
- Kovach, A.R.; Hoff, K.E.; Canty, J.T.; Orans, J.; Brennan, R.G. Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA 2014, 20, 1548–1559. [Google Scholar] [CrossRef] [Green Version]
- Stanek, K.A.; Patterson-West, J.; Randolph, P.S.; Mura, C. Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: Conservation of the lateral RNA-binding mode. Acta Crystallogr. D Struct. Biol. 2017, 73, 294–315. [Google Scholar] [CrossRef] [Green Version]
- Schulz, E.C.; Seiler, M.; Zuliani, C.; Voigt, F.; Rybin, V.; Pogenberg, V.; Mucke, N.; Wilmanns, M.; Gibson, T.J.; Barabas, O. Intermolecular base stacking mediates RNA-RNA interaction in a crystal structure of the RNA chaperone Hfq. Sci. Rep. 2017, 7, 9903. [Google Scholar] [CrossRef] [Green Version]
- Horstmann, N.; Orans, J.; Valentin-Hansen, P.; Shelburne, S.A., 3rd; Brennan, R.G. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012, 40, 11023–11035. [Google Scholar] [CrossRef] [Green Version]
- Wen, B.; Wang, W.; Zhang, J.; Gong, Q.; Shi, Y.; Wu, J.; Zhang, Z. Structural and dynamic properties of the C-terminal region of the Escherichia coli RNA chaperone Hfq: Integrative experimental and computational studies. Phys. Chem. Chem. Phys. 2017, 19, 21152–21164. [Google Scholar] [CrossRef]
- Olsen, A.S.; Moller-Jensen, J.; Brennan, R.G.; Valentin-Hansen, P. C-Terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010, 404, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Vecerek, B.; Rajkowitsch, L.; Sonnleitner, E.; Schroeder, R.; Blasi, U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008, 36, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago-Frangos, A.; Kavita, K.; Schu, D.J.; Gottesman, S.; Woodson, S.A. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. USA 2016, 113, E6089–E6096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orans, J.; Kovach, A.R.; Hoff, K.E.; Horstmann, N.M.; Brennan, R.G. Crystal structure of an Escherichia coli Hfq Core (residues 2-69)-DNA complex reveals multifunctional nucleic acid binding sites. Nucleic Acids Res. 2020, 48, 3987–3997. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trepout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35, e00190. [Google Scholar] [CrossRef]
- Partouche, D.; Militello, V.; Gomesz-Zavaglia, A.; Wien, F.; Sandt, C.; Arluison, V. In situ characterization of Hfq bacterial amyloid: A Fourier-transform infrared spectroscopy study. Pathogens 2019, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.; Debelouchina, G.T.; Bayro, M.J.; Clare, D.K.; Caporini, M.A.; Bajaj, V.S.; Jaroniec, C.P.; Wang, L.; Ladizhansky, V.; Muller, S.A.; et al. Atomic structure and hierarchical assembly of a cross-beta amyloid fibril. Proc. Natl. Acad. Sci. USA 2013, 110, 5468–5473. [Google Scholar] [CrossRef] [Green Version]
- Gremer, L.; Scholzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Ju, Y.; Tam, K.Y. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen. Res. 2022, 17, 543–549. [Google Scholar]
- Maury, C.P. The emerging concept of functional amyloid. J. Intern. Med. 2009, 265, 329–334. [Google Scholar] [CrossRef]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl. Acad. Sci. USA 2007, 104, 17388–17393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khambhati, K.; Patel, J.; Saxena, V.; A, P.; Jain, N. Gene Regulation of Biofilm-Associated Functional Amyloids. Pathogens 2021, 10, 490. [Google Scholar] [CrossRef]
- Ragonis-Bachar, P.; Landau, M. Functional and pathological amyloid structures in the eyes of 2020 cryo-EM. Curr. Opin. Struct. Biol. 2021, 68, 184–193. [Google Scholar] [CrossRef]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar] [CrossRef] [Green Version]
- Sledjeski, D.; Gottesman, S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar] [CrossRef] [Green Version]
- Lease, R.A.; Cusick, M.E.; Belfort, M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 1998, 95, 12456–12461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayrol, B.; Fortas, E.; Martret, C.; Cech, G.; Kloska, A.; Caulet, S.; Barbet, M.; Trepout, S.; Marco, S.; Taghbalout, A.; et al. Riboregulation of the bacterial actin-homolog MreB by DsrA small noncoding RNA. Integr. Biol. 2015, 7, 128–141. [Google Scholar] [CrossRef]
- Lalaouna, D.; Morissette, A.; Carrier, M.C.; Masse, E. DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli. Mol. Microbiol. 2015, 98, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Liu, X.; Yang, L.; Sun, Y.; Gong, Q.; Wu, J.; Shi, Y. The important conformational plasticity of DsrA sRNA for adapting multiple target regulation. Nucleic Acids Res. 2017, 45, 9625–9639. [Google Scholar] [CrossRef] [Green Version]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-Mediated General Stress Response in Escherichia coli (*). Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [Green Version]
- Majdalani, N.; Vanderpool, C.K.; Gottesman, S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 93–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lease, R.A.; Woodson, S.A. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004, 344, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Soper, T.J.; Woodson, S.A. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 2008, 14, 1907–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef] [PubMed]
- Wien, F.; Geinguenaud, F.; Grange, W.; Arluison, V. SRCD and FTIR Spectroscopies to Monitor Protein-Induced Nucleic Acid Remodeling. Methods Mol. Biol. 2021, 2209, 87–108. [Google Scholar]
- Malabirade, A.; Morgado-Brajones, J.; Trepout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 10724. [Google Scholar] [CrossRef]
- Wien, F.; Martinez, D.; Le Brun, E.; Jones, N.C.; Vronning Hoffmann, S.; Waeytens, J.; Berbon, M.; Habenstein, B.; Arluison, V. The Bacterial Amyloid-Like Hfq Promotes In Vitro DNA Alignment. Microorganisms 2019, 7, 639. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef] [Green Version]
- Miles, A.J.; Wallace, B.A. CDtoolX, A downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef]
- Le Brun, E.; Arluison, V.; Wien, F. Application of Synchrotron Radiation Circular Dichroism for RNA Structural Analysis. Methods Mol. Biol. 2020, 2113, 135–148. [Google Scholar]
- Repoila, F.; Gottesman, S. Temperature sensing by the dsrA promoter. J. Bacteriol. 2003, 185, 6609–6614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, D.S.; Williams, A.L., Jr. CD of nucleic acids: III. Calculated CD of RNAs from new A, U, G, and C transition-moment parameters. Biopolymers 1986, 25, 1461–1491. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.I.S.; Nielsen, L.M.; Hoffmann, S.V.; Nielsen, S.B. Vacuum-ultraviolet circular dichroism spectroscopy of DNA: A valuable tool to elucidate topology and electronic coupling in DNA. Phys. Chem. Chem. Phys. 2010, 12, 9581–9596. [Google Scholar] [CrossRef]
- Heus, H.A.; Pardi, A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 1991, 253, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Varani, G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 379–404. [Google Scholar] [CrossRef]
- Liu, X.; Shen, S.; Wu, P.; Li, F.; Liu, X.; Wang, C.; Gong, Q.; Wu, J.; Yao, X.; Zhang, H.; et al. Structural insights into dimethylation of 12S rRNA by TFB1M: Indispensable role in translation of mitochondrial genes and mitochondrial function. Nucleic Acids Res. 2019, 47, 7648–7665. [Google Scholar] [CrossRef] [Green Version]
- Jucker, F.M.; Heus, H.A.; Yip, P.F.; Moors, E.H.; Pardi, A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996, 264, 968–980. [Google Scholar] [CrossRef]
- Cordeiro, Y.; Macedo, B.; Silva, J.L.; Gomes, M.P.B. Pathological implications of nucleic acid interactions with proteins associated with neurodegenerative diseases. Biophys. Rev. 2014, 6, 97–110. [Google Scholar] [CrossRef] [Green Version]
- Greco, S.; Zaccagnini, G.; Fuschi, P.; Voellenkle, C.; Carrara, M.; Sadeghi, I.; Bearzi, C.; Maimone, B.; Castelvecchio, S.; Stellos, K.; et al. Increased BACE1-AS long noncoding RNA and beta-amyloid levels in heart failure. Cardiovasc. Res. 2017, 113, 453–463. [Google Scholar] [CrossRef]
- Chakraborty, P.; Riviere, G.; Liu, S.; de Opakua, A.I.; Dervisoglu, R.; Hebestreit, A.; Andreas, L.B.; Vorberg, I.M.; Zweckstetter, M. Co-factor-free aggregation of tau into seeding-competent RNA-sequestering amyloid fibrils. Nat. Commun. 2021, 12, 4231. [Google Scholar] [CrossRef]
- Wang, M.; Tao, X.; Jacob, M.D.; Bennett, C.A.; Ho, J.J.D.; Gonzalgo, M.L.; Audas, T.E.; Lee, S. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018, 24, 1713–1721.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Sample | Tm at ~180 nm (°C) |
|---|---|
| DsrAcore + rpoSreg | 33.6 ± 2.2 |
| DsrAcore + rpoSreg + CTR | 36.6 ± 0.8 |
| DsrAcore + rpoSreg + Hfq | 37.0 ± 0.9 |
| DsrAcore:rpoSreg | 39.9 ± 0.4 |
| DsrAcore:rpoSreg + CTR | 44.2 ± 0.7 |
| DsrAcore | 42.1 ± 0.3 |
| DsrAcore + CTR | 45.5 ± 0.8 |
| DsrAcoremut | 49.1 ± 1.2 |
| DsrAcoremut + CTR | 50.3 ± 0.9 |
| rpoSrbs + rpoSreg | 29.5 ± 2.2 |
| rpoSrbs + rpoSreg + CTR | 31.6 ± 1.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turbant, F.; Wu, P.; Wien, F.; Arluison, V. The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology 2021, 10, 900. https://doi.org/10.3390/biology10090900
Turbant F, Wu P, Wien F, Arluison V. The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology. 2021; 10(9):900. https://doi.org/10.3390/biology10090900
Chicago/Turabian StyleTurbant, Florian, Pengzhi Wu, Frank Wien, and Véronique Arluison. 2021. "The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing" Biology 10, no. 9: 900. https://doi.org/10.3390/biology10090900
APA StyleTurbant, F., Wu, P., Wien, F., & Arluison, V. (2021). The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology, 10(9), 900. https://doi.org/10.3390/biology10090900