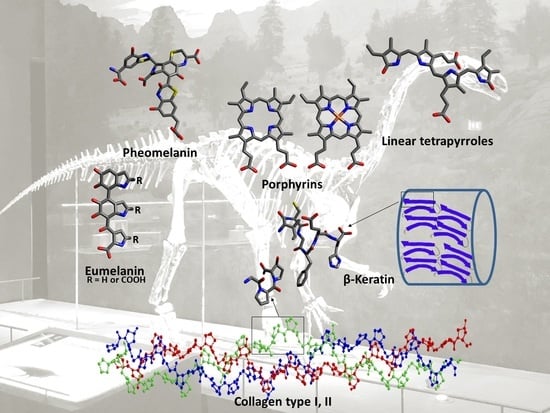
Chemistry and Analysis of Organic Compounds in Dinosaurs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Analytical Techniques to Investigate Preserved Organic Compounds
2.1. Microscopy
2.2. Spectroscopy and Spectrometry
2.2.1. UV/Vis Spectroscopy
2.2.2. Infrared and Raman Spectroscopy
2.2.3. Mass Spectrometry
2.3. Immunological Techniques
3. Organic Compounds Found in Dinosaurs
3.1. Pigments
3.1.1. Porphyrins
3.1.2. Melanins
3.2. Proteins
3.2.1. Collagens
Collagen Type I
Collagen Type II
3.2.2. Keratins
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATR-IR | Attenuated-total reflection infrared spectroscopy |
| BN-PAGE | Blue native-polyacrylamide gel electrophoresis |
| EDS | Energy-dispersive X-ray spectrometry |
| ELISA | Enzyme linked immunosorbent assay |
| ESI-MS | Electrospray ionization mass spectrometry |
| FESEM | Field emission scanning electron microscopy |
| FT-ICR-MS | Fourier transform ion cyclotron resonance mass spectrometry |
| FTIR | Fourier transform infrared spectroscopy |
| IR | Infrared spectroscopy |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| NMR | Nuclear magnetic resonance |
| OM | Optical microscopy |
| Py-GC-MS | Pyrolysis gas-chromatography mass spectrometry |
| q/TOF-MS | Quadrupole time-of-flight mass spectrometry |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SEM | Scanning electron microscopy |
| SR-FTIR | Synchrotron-radiation Fourier transform infrared spectroscopy |
| TEM | Transmission electron microscopy |
| TOF-SIMS | Time-of-flight secondary-ion mass spectrometry |
| UV/VIS | Ultraviolet-visible light |
| VPSEM | Variable-pressure scanning electron microscopy |
References
- Eglinton, G.; Logan, G.A. Molecular preservation. Philos. Trans. R. Soc. 1991, 333, 315–328. [Google Scholar] [CrossRef]
- Schweitzer, M.H. Soft tissue preservation in terrestrial Mesozoic vertebrates. Annu. Rev. Earth Planet. Sci. 2011, 39, 187–216. [Google Scholar] [CrossRef]
- Lee, Y.C.; Chiang, C.C.; Huang, P.Y.; Chung, C.Y.; Huang, T.D.; Wang, C.C.; Chen, C.I.; Chang, R.S.; Liao, C.H.; Reisz, R.R. Evidence of preserved collagen in an Early Jurassic sauropodomorph dinosaur revealed by synchrotron FTIR microspectroscopy. Nat. Commun. 2017, 8, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Luo, Z.-X.; Yuan, C.-X.; Wible, J.R.; Zhang, J.-P.; Georgi, J.A. The earliest known eutherian mammal. Nature 2002, 416, 816–822. [Google Scholar] [CrossRef]
- Gioncada, A.; Collareta, A.; Gariboldi, K.; Lambert, O.; Di Celma, C.; Bonaccorsi, E.; Urbina, M.; Bianucci, G. Inside baleen: Exceptional microstructure preservation in a late Miocene whale skeleton from Peru. Geology 2016, 44, 839–842. [Google Scholar] [CrossRef]
- Cadena, E.-A. In situ SEM/EDS compositional characterization of osteocytes and blood vessels in fossil and extant turtles on untreated bone surfaces: Different preservational pathways microns away. PeerJ 2020, 8, e9833. [Google Scholar] [CrossRef]
- Surmik, D.; Dulski, M.; Kremer, B.; Szade, J.; Pawlicki, R. Iron-mediated deep-time preservation of osteocytes in a Middle Triassic reptile bone. Hist. Biol. 2019, 33, 186–193. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Zheng, W.; Cleland, T.P.; Goodwin, M.B.; Boatman, E.; Theil, E.; Marcus, M.A.; Fakra, S.C. A role for iron and oxygen chemistry in preserving soft tissues, cells and molecules from deep time. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132741. [Google Scholar] [CrossRef] [Green Version]
- Wiemann, J.; Fabbri, M.; Yang, T.R.; Stein, K.; Sander, P.M.; Norell, M.A.; Briggs, D.E.G. Fossilization transforms vertebrate hard tissue proteins into N-heterocyclic polymers. Nat. Commun. 2018, 9, 4741. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, M.H.; Wittmeyer, J.L.; Horner, J.R. Soft tissue and cellular preservation in vertebrate skeletal elements from the Cretaceous to the present. Proc. R. Soc. B Biol. Sci. 2007, 274, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Briggs, D.E.G. Molecular taphonomy of animal and plant cuticles: Selective preservation and diagenesis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1999, 354, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.J.; Nielsen-Marsh, C.M.; Hiller, J.; Smith, C.I.; Roberts, J.P.; Prigodich, R.V.; Wess, T.J.; Csapò, J.; Millard, A.R.; Turner-Walker, G. The survival of organic matter in bone: A review. Archaeometry 2002, 44, 383–394. [Google Scholar] [CrossRef] [Green Version]
- Briggs, D.E.G.; Mcmahon, S. The role of experiments in investigating the taphonomy of exceptional preservation. Palaeontology 2016, 59, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Keenan, S.W. From bone to fossil: A review of the diagenesis of bioapatite. Am. Mineral. 2016, 101, 1943–1951. [Google Scholar] [CrossRef]
- Butterfield, N.J. Exceptional fossil preservation and the Cambrian Explosion. Integr. Comp. Biol. 2003, 43, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Pawlicki, R.; Korbel, A.; Kubiak, H. Cells, collagen fibrils and vessels in dinosaur bone. Nature 1966, 211, 655–657. [Google Scholar] [CrossRef]
- Zhang, F.; Kearns, S.L.; Orr, P.J.; Benton, M.J.; Zhou, Z.; Johnson, D.; Xu, X.; Wang, X. Fossilized melanosomes and the colour of Cretaceous dinosaurs and birds. Nature 2010, 463, 1075–1078. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Marshall, M.; Carron, K.; Bohle, D.S.; Busse, S.C.; Arnold, E.V.; Barnard, D.; Horner, J.R.; Starkey, J.R. Heme compounds in dinosaur trabecular bone. Proc. Natl. Acad. Sci. USA 1997, 94, 6291–6296. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, M.H.; Suo, Z.; Avci, R.; Asara, J.M.; Allen, M.A.; Arce, F.T.; Horner, J.R. Analyses of soft tissue from Tyrannosaurus rex suggest the presence of protein. Science 2007, 316, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Asara, J.M.; Schweitzer, M.H.; Freimark, L.M.; Phillips, M.; Cantley, L.C. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science 2007, 316, 280–285. [Google Scholar] [CrossRef]
- Wiemann, J.; Yang, T.R.; Norell, M.A. Dinosaur egg colour had a single evolutionary origin. Nature 2018, 563, 555–558. [Google Scholar] [CrossRef]
- Wiemann, J.; Yang, T.R.; Sander, P.N.; Schneider, M.; Engeser, M.; Kath-Schorr, S.; Müller, C.E.; Sander, P.M. Dinosaur origin of egg color: Oviraptors laid blue-green eggs. PeerJ 2017, 5, e3706. [Google Scholar] [CrossRef] [Green Version]
- Moyer, A.E.; Zheng, W.; Schweitzer, M.H. Microscopic and immunohistochemical analyses of the claw of the nesting dinosaur, Citipati osmolskae. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161997. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, M.H.; Watt, J.A.; Avci, R.; Knapp, L.; Chiappe, L.; Norell, M.; Marshall, M. Beta-keratin specific immunological reactivity in feather-like structures of the cretaceous alvarezsaurid, Shuvuuia deserti. J. Exp. Zool. 1999, 285, 146–157. [Google Scholar] [CrossRef]
- Lindgren, J.; Sjövall, P.; Carney, R.M.; Cincotta, A.; Uvdal, P.; Hutcheson, S.W.; Gustafsson, O.; Lefèvre, U.; Escuillié, F.; Heimdal, J.; et al. Molecular composition and ultrastructure of Jurassic paravian feathers. Sci. Rep. 2015, 5, 13520. [Google Scholar] [CrossRef] [Green Version]
- Bertazzo, S.; Maidment, S.C.R.; Kallepitis, C.; Fearn, S.; Stevens, M.M.; Xie, H.N. Fibres and cellular structures preserved in 75-million-year-old dinosaur specimens. Nat. Commun. 2015, 6, 7352. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.M.; Henderson, D.M.; Vinther, J.; Fletcher, I.; Sistiaga, A.; Herrera, J.; Summons, R.E. An exceptionally preserved three-dimensional armored dinosaur reveals insights into coloration and Cretaceous predator-prey dynamics. Curr. Biol. 2017, 27, 2514–2521.e3. [Google Scholar] [CrossRef] [Green Version]
- Lingham-Soliar, T.; Plodowski, G. The integument of Psittacosaurus from Liaoning Province, China: Taphonomy, epidermal patterns and color of a ceratopsian dinosaur. Naturwissenschaften 2010, 97, 479–486. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Zheng, W.; Organ, C.L.; Avci, R.; Suo, Z.; Freimark, L.M.; Lebleu, V.S.; Duncan, M.B.; Heiden, M.G.V.; Neveu, J.M.; et al. Biomolecular characterization and protein sequences of the Campanian hadrosaur B. canadensis. Science 2009, 324, 626–631. [Google Scholar] [CrossRef]
- Schroeter, E.R.; Dehart, C.J.; Cleland, T.P.; Zheng, W.; Thomas, P.M.; Kelleher, N.L.; Bern, M.; Schweitzer, M.H. Expansion for the Brachylophosaurus canadensis collagen I sequence and additional evidence of the preservation of Cretaceous protein. J. Proteome Res. 2017, 16, 920–932. [Google Scholar] [CrossRef] [Green Version]
- Bailleul, A.M.; Zheng, W.; Horner, J.R.; Hall, B.K.; Holliday, C.M.; Schweitzer, M.H. Evidence of proteins, chromosomes and chemical markers of DNA in exceptionally preserved dinosaur cartilage. Natl. Sci. Rev. 2020, 7, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Hu, L.; Zhao, T. Applications of chemical imaging techniques in paleontology. Natl. Sci. Rev. 2019, 6, 1040–1053. [Google Scholar] [CrossRef]
- Cleland, T.P.; Schroeter, E.R. A comparison of common mass spectrometry approaches for paleoproteomics. J. Proteome Res. 2018, 17, 936–945. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Schroeter, E.R.; Cleland, T.P.; Zheng, W. Paleoproteomics of Mesozoic dinosaurs and other Mesozoic fossils. Proteomics 2019, 19, 1800251. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Avci, R.; Collier, T.; Goodwin, M.B. Microscopic, chemical and molecular methods for examining fossil preservation. Comptes Rendus Palevol 2008, 7, 159–184. [Google Scholar] [CrossRef]
- Zhou, W.; Apkarian, R.; Wang, Z.L.; Joy, D. Fundamentals of scanning electron microscopy (SEM). In Scanning Microscopy for Nanotechnology; Springer: New York, NY, USA, 2006; pp. 1–40. [Google Scholar]
- Adams, F.; Barbante, C. Electron-based imaging techniques. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2015; Volume 69, pp. 269–313. [Google Scholar]
- Saitta, E.T.; Liang, R.; Lau, M.C.; Brown, C.M.; Longrich, N.R.; Kaye, T.G.; Novak, B.J.; Salzberg, S.L.; Norell, M.A.; Abbott, G.D.; et al. Cretaceous dinosaur bone contains recent organic material and provides an environment conducive to microbial communities. Elife 2019, 8, e46205. [Google Scholar] [CrossRef]
- Relucenti, M.; Familiari, G.; Donfrancesco, O.; Taurino, M.; Li, X.; Chen, R.; Artini, M.; Papa, R.; Selan, L. Microscopy methods for biofilm imaging: Focus on SEM and VP-SEM pros and cons. Biology 2021, 10, 51. [Google Scholar] [CrossRef]
- Klijn, M.E.; Hubbuch, J. Application of ultraviolet, visible, and infrared light imaging in protein-based biopharmaceutical formulation characterization and development studies. Eur. J. Pharm. Biopharm. 2021, 165, 319–336. [Google Scholar] [CrossRef]
- Picollo, M.; Aceto, M.; Vitorino, T. UV-Vis spectroscopy. Phys. Sci. Rev. 2019, 4, 20180008. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rehman, K. Ultraviolet-visible (UV-VIS) spectroscopy. In Essentials of Pharmaceutical Analysis; Springer: Singapore, 2020; pp. 29–56. ISBN 978-981-15-1547-7. [Google Scholar]
- Olcott Marshall, A.; Marshall, C.P. Vibrational spectroscopy of fossils. Palaeontology 2015, 58, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Loutherback, K.; Birarda, G.; Chen, L.; N Holman, H.-Y. Microfluidic approaches to synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectral microscopy of living biosystems. Protein Pept. Lett. 2016, 23, 273–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Lu, X.; Yin, X.; Tong, Y.; Peng, W.; Wu, L.; Li, H.; Yang, Y.; Gu, J.; Xiao, T.; et al. Synchrotron radiation-based Fourier-transform infrared spectromicroscopy for characterization of the protein/peptide distribution in single microspheres. Acta Pharm. Sin. B 2015, 5, 270–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baeten, V.; Dardenne, P. Spectroscopy: Developments in instrumentation and analysis. Grasas Aceites 2002, 53, 45–63. [Google Scholar] [CrossRef]
- Lohumi, S.; Kim, M.S.; Qin, J.; Cho, B.-K. Raman imaging from microscopy to macroscopy: Quality and safety control of biological materials. TrAC Trends Anal. Chem. 2017, 93, 183–198. [Google Scholar] [CrossRef]
- Raman, C.V.; Krishnan, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Marigheto, N.A.; Kemsley, E.K.; Potter, J.; Belton, P.S.; Wilson, R.H. Effects of sample heating in FT-Raman spectra of biological materials. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 1996, 52, 1571–1579. [Google Scholar] [CrossRef]
- Milman, B.L. General principles of identification by mass spectrometry. TrAC Trends Anal. Chem. 2015, 69, 24–33. [Google Scholar] [CrossRef]
- Altelaar, A.F.M.; Luxembourg, S.L.; McDonnell, L.A.; Piersma, S.R.; Heeren, R.M.A. Imaging mass spectrometry at cellular length scales. Nat. Protoc. 2007, 2, 1185–1196. [Google Scholar] [CrossRef]
- Thiel, V.; Sjövall, P. Using time-of-flight secondary ion mass spectrometry to study Biomarkers. Annu. Rev. Earth Planet. Sci. 2011, 39, 125–156. [Google Scholar] [CrossRef]
- Greenwalt, D.E.; Goreva, Y.S.; Siljeström, S.M.; Rose, T.; Harbach, R.E. Hemoglobin-derived porphyrins preserved in a Middle Eocene blood-engorged mosquito. Proc. Natl. Acad. Sci. USA 2013, 110, 18496–18500. [Google Scholar] [CrossRef] [Green Version]
- Lindgren, J.; Uvdal, P.; Sjövall, P.; Nilsson, D.E.; Engdahl, A.; Schultz, B.P.; Thiel, V. Molecular preservation of the pigment melanin in fossil melanosomes. Nat. Commun. 2012, 3, 824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winograd, N. The Magic of Cluster SIMS. Anal. Chem. 2005, 77, 142 A–149 A. [Google Scholar] [CrossRef] [Green Version]
- Kozole, J.; Winograd, N. Cluster secondary ion mass spectrometry. In Surface Analysis and Techniques in Biology; Smentkowski, V.S., Ed.; Springer: Cham, Switzerland, 2014; pp. 71–98. [Google Scholar]
- Meier, D.; Faix, O. Pyrolysis-gas chromatography-mass spectrometry. In Methods in Lignin Chemistry; Lin, S.Y., Dence, C.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 177–199. ISBN 13:978-3-642-74065-7. [Google Scholar]
- Stankiewicz, B.A.; Briggs, D.E.G.; Evershed, R.P.; Flannery, M.B.; Wuttke, M. Preservation of chitin in 25-million-year-old fossils. Science 1997, 276, 1541–1543. [Google Scholar] [CrossRef]
- Stankiewicz, B.A.; Poinar, H.N.; Briggs, D.E.G.; Evershed, R.P.; Poinar, J. Chemical preservation of plants and insects in natural resins. Proc. R. Soc. B Biol. Sci. 1998, 265, 641–647. [Google Scholar] [CrossRef] [Green Version]
- Stankiewicz, B.A.; Mastalerz, M.; Kruge, M.A.; Van Bergen, P.F.; Sadowska, A. A comparative study of modern and fossil cone scales and seeds of conifers: A geochemical approach. New Phytol. 1997, 135, 375–393. [Google Scholar] [CrossRef]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef]
- El-Aneed, A.; Cohen, A.; Banoub, J. Mass spectrometry, review of the basics: Electrospray, MALDI, and commonly used mass analyzers. Appl. Spectrosc. Rev. 2009, 44, 210–230. [Google Scholar] [CrossRef]
- Crowther, J.R. Overview of ELISA in relation to other disciplines. In The ELISA Guidebook. Methods in Molecular Biology; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2009; pp. 1–8. ISBN 978-1-60327-254-4. [Google Scholar]
- Yu, H.-W.; Halonen, M.J.; Pepper, I.L. Immunological methods. In Environmental Microbiology; Pepper, I.L., Gerba, C.P., Gentry, T.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 245–269. ISBN 978-0-12-394626-3. [Google Scholar]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Stanker, L.H.; Hnasko, R.M. A double-sandwich ELISA for identification of monoclonal antibodies suitable for sandwich immunoassays. In Methods in Molecular Biology; Hnasko, R., Ed.; Humana Press: New York, NY, USA, 2015; pp. 69–78. ISBN 978-1-4939-2741-8. [Google Scholar]
- Gan, S.D.; Patel, K.R. Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Investig. Dermatol. 2013, 133, e12. [Google Scholar] [CrossRef] [Green Version]
- Makarananda, K.; Weir, L.R.; Neal, G.E. Competitive ELISA. In Immunochemical Protocols; Pound, J.D., Ed.; Humana Press: Totowa, NJ, USA, 1998; pp. 155–160. ISBN 978-1-59259-257-9. [Google Scholar]
- Gallagher, S.R. SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Curr. Protoc. Essent. Lab. Tech. 2008, 6, 7-3. [Google Scholar] [CrossRef]
- Yang, P.-C.; Mahmood, T. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429. [Google Scholar] [CrossRef]
- Crichton, P.G.; Harding, M.; Ruprecht, J.J.; Lee, Y.; Kunji, E.R.S. Lipid, detergent, and Coomassie Blue G-250 affect the migration of small membrane proteins in Blue Native gels. J. Biol. Chem. 2013, 288, 22163–22173. [Google Scholar] [CrossRef] [Green Version]
- Na Ayutthaya, P.P.; Lundberg, D.; Weigel, D.; Li, L. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the analysis of protein oligomers in plants. Curr. Protoc. Plant Biol. 2020, 5, e20107. [Google Scholar] [CrossRef]
- Gillett, C.E. Immunohistochemistry. In Breast Cancer Research Protocols; Brooks, S.A., Harris, A.L., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 191–200. [Google Scholar]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An introduction to the performance of immunohistochemistry. In Biobanking: Methods and Protocols; Yong, W.H., Ed.; Springer: New York, NY, USA, 2019; pp. 289–298. ISBN 978-1-4939-8935-5. [Google Scholar]
- D’Amico, F.; Skarmoutsou, E.; Stivala, F. State of the art in antigen retrieval for immunohistochemistry. J. Immunol. Methods 2009, 341, 1–18. [Google Scholar] [CrossRef]
- Alfonso-Rojas, A.; Cadena, E.-A. Exceptionally preserved ‘skin’ in an Early Cretaceous fish from Colombia. PeerJ 2020, 8, e9479. [Google Scholar] [CrossRef]
- Roy, A.; Pittman, M.; Saitta, E.T.; Kaye, T.G.; Xu, X. Recent advances in amniote palaeocolour reconstruction and a framework for future research. Biol. Rev. 2020, 95, 22–50. [Google Scholar] [CrossRef] [Green Version]
- Vinther, J. A guide to the field of palaeo colour. BioEssays 2015, 37, 643–656. [Google Scholar] [CrossRef]
- Vinther, J. Reconstructing vertebrate paleocolor. Annu. Rev. Earth Planet. Sci. 2020, 48, 345–375. [Google Scholar] [CrossRef]
- Gueneli, N.; Mckenna, A.M.; Ohkouchi, N.; Boreham, C.J.; Beghin, J.; Javaux, E.J.; Brocks, J.J. 1.1-billion-year-old porphyrins establish a marine ecosystem dominated by bacterial primary producers. Proc. Natl. Acad. Sci. USA 2018, 115, E6978–E6986. [Google Scholar] [CrossRef] [Green Version]
- Tahoun, M.; Gee, C.T.; McCoy, V.E.; Sander, P.M.; Müller, C.E. Chemistry of porphyrins in fossil plants and animals. RSC Adv. 2021, 11, 7552–7563. [Google Scholar] [CrossRef]
- Asher, S.A. Resonance Raman spectroscopy of hemoglobin. In Methods in Enzymology; Antonini, E., Chiancone, E., Rossi-Bernardi, L., Eds.; Academic Press: New York, NY, USA, 1981; pp. 371–413. [Google Scholar]
- Alleon, J.; Montagnac, G.; Reynard, B.; Brulé, T.; Thoury, M.; Gueriau, P. Pushing Raman spectroscopy over the edge: Purported signatures of organic molecules in fossil animals are instrumental artefacts. BioEssays 2021, 43, 2000295. [Google Scholar] [CrossRef]
- Wiemann, J.; Briggs, D.E.G. Raman spectroscopy is a powerful tool in molecular paleobiology: An analytical response to Alleon et al. (https://doi.org/10.1002/bies.202000295). BioEssays 2022, 44, 2100070. [Google Scholar] [CrossRef]
- Nasti, T.H.; Timares, L. MC1R, Eumelanin and Pheomelanin: Their role in determining the susceptibility to skin cancer. Photochem. Photobiol. 2015, 91, 188–200. [Google Scholar] [CrossRef] [Green Version]
- Suzukawa, A.A.; Vieira, A.; Winnischofer, S.M.B.; Scalfo, A.C.; Di Mascio, P.; Ferreira, A.M.D.C.; Ravanat, J.-L.; Martins, D.D.L.; Rocha, M.E.M.; Martinez, G.R. Novel properties of melanins include promotion of DNA strand breaks, impairment of repair, and reduced ability to damage DNA after quenching of singlet oxygen. Free Radic. Biol. Med. 2012, 52, 1945–1953. [Google Scholar] [CrossRef]
- Lindgren, J.; Moyer, A.; Schweitzer, M.H.; Sjövall, P.; Uvdal, P.; Nilsson, D.E.; Heimdal, J.; Engdahl, A.; Gren, J.A.; Schultz, B.P.; et al. Interpreting melanin-based coloration through deep time: A critical review. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150614. [Google Scholar] [CrossRef]
- Ortonne, J.-P. Photoprotective properties of skin melanin. Br. J. Dermatol. 2002, 146, 7–10. [Google Scholar] [CrossRef]
- D’Ischia, M.; Napolitano, A.; Ball, V.; Chen, C.-T.; Buehler, M.J. Polydopamine and Eumelanin: From structure–property relationships to a unified tailoring strategy. Acc. Chem. Res. 2014, 47, 3541–3550. [Google Scholar] [CrossRef]
- Pezzella, A.; Napolitano, A.; D’Ischia, M.; Prota, G.; Seraglia, R.; Traldi, P. Identification of partially degraded oligomers of 5,6-dihydroxyindole-2-carboxylic acid in sepia melanin by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1997, 11, 368–372. [Google Scholar] [CrossRef]
- Falk, H.; Wolkenstein, K. Natural product molecular fossils. In Progress in the Chemistry of Organic Natural Products; Kinghorn, A.D., Falk, H., Gibbons, S., Kobayashi, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–126. ISBN 978-3-319-45618-8. [Google Scholar]
- Cao, W.; Zhou, X.; McCallum, N.C.; Hu, Z.; Ni, Q.Z.; Kapoor, U.; Heil, C.M.; Cay, K.S.; Zand, T.; Mantanona, A.J.; et al. Unraveling the structure and function of melanin through synthesis. J. Am. Chem. Soc. 2021, 143, 2622–2637. [Google Scholar] [CrossRef]
- Lindgren, J.; Sjövall, P.; Carney, R.M.; Uvdal, P.; Gren, J.A.; Dyke, G.; Schultz, B.P.; Shawkey, M.D.; Barnes, K.R.; Polcyn, M.J. Skin pigmentation provides evidence of convergent melanism in extinct marine reptiles. Nature 2014, 506, 484–488. [Google Scholar] [CrossRef]
- Colleary, C.; Dolocan, A.; Gardner, J.; Singh, S.; Wuttke, M.; Rabenstein, R.; Habersetzer, J.; Schaal, S.; Feseha, M.; Clemens, M.; et al. Chemical, experimental, and morphological evidence for diagenetically altered melanin in exceptionally preserved fossils. Proc. Natl. Acad. Sci. USA 2015, 112, 12592–12597. [Google Scholar] [CrossRef] [Green Version]
- Moyer, A.E.; Zheng, W.; Johnson, E.A.; Lamanna, M.C.; Li, D.; Lacovara, K.J.; Schweitzer, M.H. Melanosomes or microbes: Testing an alternative hypothesis for the origin of microbodies in fossil feathers. Sci. Rep. 2015, 4, 4233. [Google Scholar] [CrossRef] [Green Version]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Härtner, T.; Straub, K.L.; Kannenberg, E. Occurrence of hopanoid lipids in anaerobic Geobacter species. FEMS Microbiol. Lett. 2005, 243, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Belin, B.J.; Busset, N.; Giraud, E.; Molinaro, A.; Silipo, A.; Newman, D.K. Hopanoid lipids: From membranes to plant–bacteria interactions. Nat. Rev. Microbiol. 2018, 16, 304–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, K.; Ito, S.; Wilby, P.R.; Sota, T.; Nakamura, A.; Bowers, C.R.; Vinther, J.; Dutta, S.; Summons, R.; Briggs, D.E.G.; et al. Direct chemical evidence for eumelanin pigment from the Jurassic period. Proc. Natl. Acad. Sci. USA 2012, 109, 10218–10223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, K.; Ito, S.; Wilby, P.R.; Sota, T.; Nakamura, A.; Russell Bowers, C.; Miller, K.E.; Dutta, S.; Summons, R.E.; Briggs, D.E.G.; et al. Impact of diagenesis and maturation on the survival of eumelanin in the fossil record. Org. Geochem. 2013, 64, 29–37. [Google Scholar] [CrossRef]
- Smejkal, G.B.; Schweitzer, M.H. Will current technologies enable dinosaur proteomics? Expert Rev. Proteom. 2007, 4, 695–699. [Google Scholar] [CrossRef] [Green Version]
- Buckley, M.; Warwood, S.; van Dongen, B.; Kitchener, A.C.; Manning, P.L. A fossil protein chimera; difficulties in discriminating dinosaur peptide sequences from modern cross-contamination. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170544. [Google Scholar] [CrossRef]
- Sawyer, R.H.; Glenn, T.; French, J.O.; Mays, B.; Shames, R.B.; Barnes, G.L.; Rhodes, W.; Ishikawa, Y. The expression of beta (β) keratins in the epidermal appendages of reptiles and birds. Am. Zool. 2000, 40, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Cell junctions, cell adhesion, and the extracellular matrix. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; pp. 1065–1126. [Google Scholar]
- Koide, T. Triple helical collagen-like peptides: Engineering and applications in matrix biology. Connect. Tissue Res. 2005, 46, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozario, T.; DeSimone, D.W. The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol. 2010, 341, 126–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorushanova, A.; Delgado, L.M.; Wu, Z.; Shologu, N.; Kshirsagar, A.; Raghunath, R.; Mullen, A.M.; Bayon, Y.; Pandit, A.; Raghunath, M.; et al. The collagen suprafamily: From biosynthesis to advanced biomaterial development. Adv. Mater. 2019, 31, 1801651. [Google Scholar] [CrossRef] [Green Version]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [Green Version]
- Curtis, R.W.; Chmielewski, J. A comparison of the collagen triple helix and coiled-coil peptide building blocks on metal ion-mediated supramolecular assembly. Pept. Sci. 2021, 113, e24190. [Google Scholar] [CrossRef]
- Yamauchi, M.; Sricholpech, M. Lysine post-translational modifications of collagen. Essays Biochem. 2012, 52, 113–133. [Google Scholar] [CrossRef] [Green Version]
- Hulmes, D.J.S. Collagen diversity, synthesis and assembly. In Collagen; Fratzl, P., Ed.; Springer: Boston, MA, USA, 2008; pp. 15–47. [Google Scholar]
- Line, S.; Rhodes, C.; Yamada, Y. Molecular Biology of Cartilage Matrix. In Cellular and Molecular Biology of Bone; Noda, M., Ed.; Academic Press: San Diego, CA, USA, 1993; pp. 539–555. [Google Scholar]
- Fedarko, N.S. Osteoblast/osteoclast development and function in osteogenesis imperfecta. In Osteogenesis Imperfecta; Shapiro, J.R., Ed.; Academic Press: San Diego, CA, USA, 2014; pp. 45–56. ISBN 9780123971654. [Google Scholar]
- Henriksen, K.; Karsdal, M.A. Type I collagen. In Biochemistry of Collagens, Laminins and Elastin; Karsdal, M.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–12. ISBN 9780128170687. [Google Scholar]
- Byers, P.H.; Bonadio, J.F. The molecular basis of clinical heterogeneity in osteogenesis imperfecta: Mutations in type I collagen genes have different effects on collagen processing. In Genetic and Metabolic Disease in Pediatrics; Lloyd, J.K., Scriver, C.R., Eds.; Butterworth & Co. (Publishers) Ltd.: Bodmin, UK, 1985; pp. 56–90. [Google Scholar]
- Bächinger, H.P.; Mizuno, K.; Vranka, J.A.; Boudko, S.P. Collagen formation and structure. In Comprehensive Natural Products II: Chemistry and Biology; Liu, H.-W., Mander, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2010; Volume 5, pp. 469–530. ISBN 9780080453828. [Google Scholar]
- Shavandi, A.; Silva, T.H.; Bekhit, A.A.; Bekhit, A.E.-D.A. Keratin: Dissolution, extraction and biomedical application. Biomater. Sci. 2017, 5, 1699–1735. [Google Scholar] [CrossRef] [Green Version]
- Bragulla, H.H.; Homberger, D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009, 214, 516–559. [Google Scholar] [CrossRef]
- Coulombe, P.A.; Omary, M.B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002, 14, 110–122. [Google Scholar] [CrossRef]
- Gu, L.-H.; Coulombe, P.A. Keratin function in skin epithelia: A broadening palette with surprising shades. Curr. Opin. Cell Biol. 2007, 19, 13–23. [Google Scholar] [CrossRef]
- Sun, T.-T.; Eichner, R.; Nelson, W.G.; Scheffer Tseng, C.G.; Weiss, R.A.; Jarvinen, M.; Woodcock-Mitchell, J. Keratin classes: Molecular markers for different types of epithelial differentiation. J. Investig. Dermatol. 1983, 81, S109–S115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korniłłowicz-Kowalska, T.; Bohacz, J. Biodegradation of keratin waste: Theory and practical aspects. Waste Manag. 2011, 31, 1689–1701. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, W.; McKittrick, J.; Meyers, M.A. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mater. Sci. 2016, 76, 229–318. [Google Scholar] [CrossRef] [Green Version]
- Fraser, R.D.B.; Parry, D.A.D. Molecular packing in the feather keratin filament. J. Struct. Biol. 2008, 162, 1–13. [Google Scholar] [CrossRef]
- Toni, M.; Dalla Valle, L.; Alibardi, L. Hard (beta-)keratins in the epidermis of reptiles: Composition, sequence, and molecular organization. J. Proteome Res. 2007, 6, 3377–3392. [Google Scholar] [CrossRef]
- Perța-Crișan, S.; Ursachi, C.Ș.; Gavrilaș, S.; Oancea, F.; Munteanu, F.-D. Closing the loop with keratin-rich fibrous materials. Polymers 2021, 13, 1896. [Google Scholar] [CrossRef]





| Organic Compound | Heme | Protoporphyrin IX | Biliverdin |
|---|---|---|---|
| Structure and exact mass |  |  |  |
| Analytical technique | HPLC-UV UV/Vis spectroscopy Raman spectroscopy | LC-ESI-q/TOF-MS Raman spectroscopy | LC-ESI-q/TOF-MS Raman spectroscopy |
| Dinosaur species and age | Tyrannosaurus rex (67 Ma) | Heyuannia huangi (66 Ma) | Heyuannia huangi (66 Ma) |
| Location of fossil | Hell Creek formation, eastern Montana, USA | Chinese provinces (Henan, Jiangxi, and Guangdong) | Chinese provinces (Henan, Jiangxi, and Guangdong) |
| Type of tissue | Extracts of trabecular bone tissues | Extract of eggshells | Extract of eggshells |
| Reference | [18] | [21,22] | [21,22] |
| Eumelanosomes and Pheomelanosomes | Eumelanin-like Pigmentation (Black and Yellow) | Eumelanin | Mixture of Pheomelanin and Eumelanin | |
|---|---|---|---|---|
| Analytical technique | SEM imaging combined with EDS | Imaging with digital camera | TOF-SIMS EDS IR micro-spectroscopy | TOF-SIMS Py-GC-MS EDS |
| Dinosaur, location and age of fossil | Sinosauropteryx (125 Ma, Dawangzhangzhi, Lingyuan City, Liaoning Province, China) Sinornithosaurus (125 Ma, Sihetun, Beipiao City, Liaoning Province, China) | Psittacosaurus (125 Ma) Yixian formation in China | Anchiornis huxleyi (150 Ma) Yaolugao locality in Jianching County, western Liaoning, China | Borealopelta markmitchelli (112 Ma) Suncor Millenium Mine, Fort McMurray, Alberta, Canada |
| Type of tissue | Integumentary filaments from the tail | Preserved epidermal scales scattered from head to tail | Filamentous epidermal appendages (“feathers”) | Integumentary structures (epidermis and keratinized scales) |
| Reference | [17] | [28] | [25] | [27] |
| Study | Collagen Type I | Analytical Technique(s) | Dinosaur Name, Location and Age | Type of Tissue | Reference |
|---|---|---|---|---|---|
| 1 | Amino acid fragments and peptide sequences (5 from α1 chain, 1 from α2 chain) | Immuno-histochemistry, ELISA, TOF-SIMS and LC-MS/MS | Tyrannosaurus rex (68 Ma) Hell Creek Formation, eastern Montana, USA | Trabecular bone | [19,20] |
| 2 | Infrared absorption bands | SR-FTIR and confocal Raman microscopy | Lufengosaurus (ca. 195 Ma) Dawa, Lufeng County, Yunnan Province, China | Rib bone (thin sections) | [3] |
| 3 | Amino acid fragments (alanine, arginine, glycine, and proline) | TOF-SIMS | Various Dinosauria (75 Ma) Dinosaur Park Formation, Alberta, Canada | Claw, ungual phalanx, astragalus, tibia, rib | [26] |
| 4 | Peptide sequences (6 for α1 chain, 2 for α2 chain) | Immuno-histochemistry, Western blot, ATR-IR, TOF-SIMS, and LC-MS/MS | Brachylophosaurus canadensis (80 Ma) Judith River Formation, eastern Montana, USA | Femur from hind limb (4 different samples) | [29] |
| 5 | Peptide sequences (6 for α1 chain, 2 for α2 chain) | Nano-LC-MS/MS and FT-ICR-MS | Brachylophosaurus canadensis (80 Ma) Judith River Formation, eastern Montana, USA | Femur from hind limb (4 different samples) | [30] |
| 6 | Collagen type II | Immunohisto-chemistry | Hypacrosaurus stebingeri (75 Ma) Two Medicine Formation, northern Montana, USA. | Calcified cartilage from supraoccipital | [31] |
| β-Keratin and Its Amino Acid Fragments | β-Keratin Epitopes | |
|---|---|---|
| Analytical technique(s) | TOF-SIMS Immunohistochemistry | Immunohistochemistry |
| Dinosaur species Location and age of fossil | Shuvuuia deserti (100 Ma) Ukhaa Tolgod in southwestern Mongolia | Citipati osmolskae (75 Ma) Djadokhta Formation of Mongolia |
| Type of tissue | Feather-like epidermal appendages | Original keratinous-like claw sheath |
| Reference | [24] | [23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahoun, M.; Engeser, M.; Namasivayam, V.; Sander, P.M.; Müller, C.E. Chemistry and Analysis of Organic Compounds in Dinosaurs. Biology 2022, 11, 670. https://doi.org/10.3390/biology11050670
Tahoun M, Engeser M, Namasivayam V, Sander PM, Müller CE. Chemistry and Analysis of Organic Compounds in Dinosaurs. Biology. 2022; 11(5):670. https://doi.org/10.3390/biology11050670
Chicago/Turabian StyleTahoun, Mariam, Marianne Engeser, Vigneshwaran Namasivayam, Paul Martin Sander, and Christa E. Müller. 2022. "Chemistry and Analysis of Organic Compounds in Dinosaurs" Biology 11, no. 5: 670. https://doi.org/10.3390/biology11050670
APA StyleTahoun, M., Engeser, M., Namasivayam, V., Sander, P. M., & Müller, C. E. (2022). Chemistry and Analysis of Organic Compounds in Dinosaurs. Biology, 11(5), 670. https://doi.org/10.3390/biology11050670