Proton Microbeam Targeted Irradiation of the Gonad Primordium Region Induces Developmental Alterations Associated with Heat Shock Responses and Cuticle Defense in Caenorhabditis elegans
Abstract
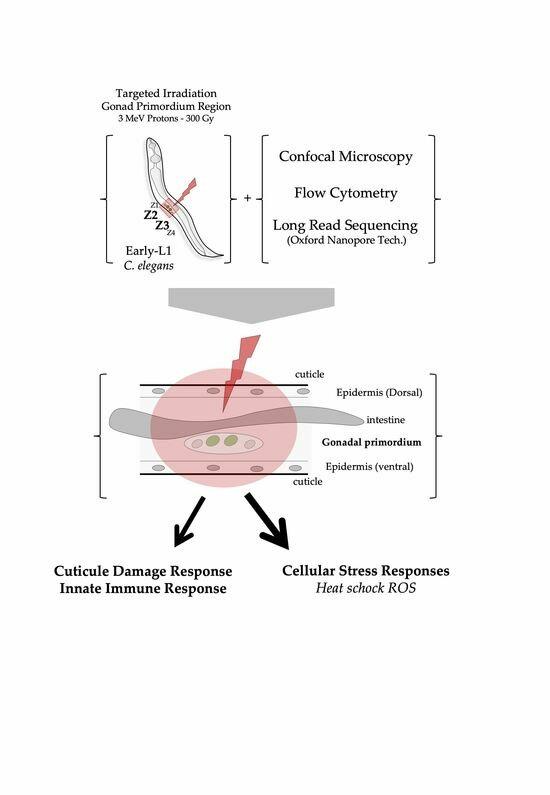
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Worm Strains and Culture
2.2. Preparation of Large Population of Synchronized C. elegans (L1 Stage)
2.3. Samples Preparation and Mounting for Irradiation
2.4. Beam Line Characteristics and Irradiation Procedures
2.5. Sample Preparation for Confocal Imaging
2.6. Nematode Population Analysis and Gene Reporter Assay Using COPAS
2.7. RNA Collection
2.8. Library Preparation and Sequencing (Oxford Nanopore Technologies, PCR-cDNA Barcoding)
2.9. Transcriptome Analysis
3. Results
3.1. Expression of the GFP::PCN-1 Fusion Protein Allows Specific Targeting under a Microbeam of Primordial Germ Region (Z2-Z3) Cells for the Monitoring of Germline and Vulva Developments
3.2. Confocal Microscopy of Radiation-Induced Alterations of Gonadal and Vulval Development in C. elegans following Selective Irradiation
3.3. High-Throughput Cell Measurement of Radiation-Induced Developmental Alterations in C. elegans following Selective and Targeted Irradiation (COPAS)
3.4. Transcriptomic Analysis Revealed Differential Expression of Genes Involved in Metabolic Stress and Innate Immune Response Related to Physical Cuticle Injuring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sulston, J.E. Neuronal Cell Lineages in the Nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 1983, 48 Pt 2, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Sulston, J.E.; Horvitz, H.R. Post-Embryonic Cell Lineages of the Nematode, Caenorhabditis elegans. Dev. Biol. 1977, 56, 110–156. [Google Scholar] [CrossRef] [PubMed]
- Kimble, J.; Hirsh, D. The Postembryonic Cell Lineages of the Hermaphrodite and Male Gonads in Caenorhabditis elegans. Dev. Biol. 1979, 70, 396–417. [Google Scholar] [CrossRef]
- Sulston, J.E.; White, J.G. Regulation and Cell Autonomy during Postembryonic Development of Caenorhabditis elegans. Dev. Biol. 1980, 78, 577–597. [Google Scholar] [CrossRef]
- Avery, L.; Horvitz, H.R. A Cell That Dies during Wild-Type C. elegans Development Can Function as a Neuron in a Ced-3 Mutant. Cell 1987, 51, 1071–1078. [Google Scholar] [CrossRef]
- Chalfie, M.; Sulston, J.E.; White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The Neural Circuit for Touch Sensitivity in Caenorhabditis elegans. J. Neurosci. 1985, 5, 956–964. [Google Scholar] [CrossRef]
- Avery, L.; Horvitz, H.R. Pharyngeal Pumping Continues after Laser Killing of the Pharyngeal Nervous System of C. elegans. Neuron 1989, 3, 473–485. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Horvitz, H.R. Control of Larval Development by Chemosensory Neurons in Caenorhabditis elegans. Science 1991, 251, 1243–1246. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Horvitz, H.R. Chemosensory Neurons with Overlapping Functions Direct Chemotaxis to Multiple Chemicals in C. elegans. Neuron 1991, 7, 729–742. [Google Scholar] [CrossRef]
- Kimble, J.E.; White, J.G. On the Control of Germ Cell Development in Caenorhabditis elegans. Dev. Biol. 1981, 81, 208–219. [Google Scholar] [CrossRef]
- Kimble, J. Alterations in Cell Lineage Following Laser Ablation of Cells in the Somatic Gonad of Caenorhabditis elegans. Dev. Biol. 1981, 87, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, H.M.; Sternberg, P.W. Multiple Cell Interactions Are Required for Fate Specification during Male Spicule Development in Caenorhabditis elegans. Development 1993, 118, 297–324. [Google Scholar] [CrossRef] [PubMed]
- Ghita, M.; Fernandez-Palomo, C.; Fukunaga, H.; Fredericia, P.M.; Schettino, G.; Bräuer-Krisch, E.; Butterworth, K.T.; McMahon, S.J.; Prise, K.M. Microbeam Evolution: From Single Cell Irradiation to Pre-Clinical Studies. Int. J. Radiat. Biol. 2018, 94, 708–718. [Google Scholar] [CrossRef]
- Muggiolu, G.; Pomorski, M.; Claverie, G.; Berthet, G.; Mer-Calfati, C.; Saada, S.; Devès, G.; Simon, M.; Seznec, H.; Barberet, P. Single α-Particle Irradiation Permits Real-Time Visualization of RNF8 Accumulation at DNA Damaged Sites. Sci. Rep. 2017, 7, 41764. [Google Scholar] [CrossRef] [PubMed]
- Muggiolu, G.; Torfeh, E.; Simon, M.; Devès, G.; Seznec, H.; Barberet, P. Recruitment Kinetics of XRCC1 and RNF8 Following MeV Proton and α-Particle Micro-Irradiation. Biology 2023, 12, 921. [Google Scholar] [CrossRef] [PubMed]
- Torfeh, E.; Simon, M.; Muggiolu, G.; Devès, G.; Vianna, F.; Bourret, S.; Incerti, S.; Barberet, P.; Seznec, H. Monte-Carlo Dosimetry and Real-Time Imaging of Targeted Irradiation Consequences in 2-Cell Stage Caenorhabditis elegans Embryo. Sci. Rep. 2019, 9, 10568. [Google Scholar] [CrossRef]
- Sleiman, A.; Lalanne, K.; Vianna, F.; Perrot, Y.; Richaud, M.; SenGupta, T.; Cardot-Martin, M.; Pedini, P.; Picard, C.; Nilsen, H.; et al. Targeted Central Nervous System Irradiation with Proton Microbeam Induces Mitochondrial Changes in Caenorhabditis elegans. Biology 2023, 12, 839. [Google Scholar] [CrossRef]
- Durante, M.; Friedl, A.A. New Challenges in Radiobiology Research with Microbeams. Radiat. Environ. Biophys. 2011, 50, 335–338. [Google Scholar] [CrossRef]
- Miller, J.H.; Chrisler, W.B.; Wang, X.; Sowa, M.B. Confocal Microscopy for Modeling Electron Microbeam Irradiation of Skin. Radiat. Environ. Biophys. 2011, 50, 365–369. [Google Scholar] [CrossRef]
- Belyakov, O.V.; Mitchell, S.A.; Parikh, D.; Randers-Pehrson, G.; Marino, S.A.; Amundson, S.A.; Geard, C.R.; Brenner, D.J. Biological Effects in Unirradiated Human Tissue Induced by Radiation Damage up to 1 Mm Away. Proc. Natl. Acad. Sci. USA 2005, 102, 14203–14208. [Google Scholar] [CrossRef]
- Zlobinskaya, O.; Girst, S.; Greubel, C.; Hable, V.; Siebenwirth, C.; Walsh, D.W.M.; Multhoff, G.; Wilkens, J.J.; Schmid, T.E.; Dollinger, G. Reduced Side Effects by Proton Microchannel Radiotherapy: Study in a Human Skin Model. Radiat. Environ. Biophys. 2013, 52, 123–133. [Google Scholar] [CrossRef]
- Sedelnikova, O.A.; Nakamura, A.; Kovalchuk, O.; Koturbash, I.; Mitchell, S.A.; Marino, S.A.; Brenner, D.J.; Bonner, W.M. DNA Double-Strand Breaks Form in Bystander Cells after Microbeam Irradiation of Three-Dimensional Human Tissue Models. Cancer Res. 2007, 67, 4295–4302. [Google Scholar] [CrossRef]
- Sugimoto, T.; Dazai, K.; Sakashita, T.; Funayama, T.; Wada, S.; Hamada, N.; Kakizaki, T.; Kobayashi, Y.; Higashitani, A. Cell Cycle Arrest and Apoptosis in Caenorhabditis elegans Germline Cells Following Heavy-Ion Microbeam Irradiation. Int. J. Radiat. Biol. 2006, 82, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Pocock, R.D.J.; Randers-Pehrson, G.; Brenner, D.J. Microbeam Irradiation of the C. elegans Nematode. J. Radiat. Res. 2009, 50 (Suppl. SA), A49–A54. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Yin, X.; Allan, R.; Lu, D.D.; Maurer, C.W.; Haimovitz-Friedman, A.; Fuks, Z.; Shaham, S.; Kolesnick, R. Ceramide Biogenesis Is Required for Radiation-Induced Apoptosis in the Germ Line of C. elegans. Science 2008, 322, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Buonanno, M.; Garty, G.; Grad, M.; Gendrel, M.; Hobert, O.; Brenner, D.J. Microbeam Irradiation of C. elegans Nematode in Microfluidic Channels. Radiat. Environ. Biophys. 2013, 52, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Sun, J.; Bian, P.; Chen, L.; Zhan, F.; Wang, J.; Xu, A.; Wang, Y.; Hei, T.K.; Wu, L. Radiation-Induced Bystander Signaling from Somatic Cells to Germ Cells in Caenorhabditis elegans. Radiat. Res. 2013, 180, 268–275. [Google Scholar] [CrossRef]
- Li, Q.; Shi, J.; Chen, L.; Zhan, F.; Yuan, H.; Wang, J.; Xu, A.; Wu, L. Spatial Function of the Oxidative DNA Damage Response in Radiation Induced Bystander Effects in Intra- and Inter-System of Caenorhabditis elegans. Oncotarget 2017, 8, 51253–51263. [Google Scholar] [CrossRef]
- Horvitz, H.R.; Sternberg, P.W. Multiple Intercellular Signalling Systems Control the Development of the Caenorhabditis elegans Vulva. Nature 1991, 351, 535–541. [Google Scholar] [CrossRef]
- Seydoux, G.; Salvage, C.; Greenwald, I. Isolation and Characterization of Mutations Causing Abnormal Eversion of the Vulva in Caenorhabditis elegans. Dev. Biol. 1993, 157, 423–436. [Google Scholar] [CrossRef]
- Stiernagle, T. Maintenance of C. elegans; WormBook: Pasadena, CA, USA, 2006; pp. 1–11. [Google Scholar] [CrossRef]
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Barberet, P.; Jouve, J.; Sorieul, S.; Alfaurt, P.; Mathieu, L. AIFIRA: A Light Ion Beam Facility for Ion Beam Analysis and Irradiation. Eur. Phys. J. Plus 2021, 136, 67. [Google Scholar] [CrossRef]
- Bourret, S.; Vianna, F.; Devès, G.; Atallah, V.; Moretto, P.; Seznec, H.; Barberet, P. Fluorescence Time-Lapse Imaging of Single Cells Targeted with a Focused Scanning Charged-Particle Microbeam. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2014, 325, 27–34. [Google Scholar] [CrossRef]
- Edelstein, A.D.; Tsuchida, M.A.; Amodaj, N.; Pinkard, H.; Vale, R.D.; Stuurman, N. Advanced Methods of Microscope Control Using μManager Software. J. Biol. Methods 2014, 1, e10. [Google Scholar] [CrossRef]
- Boyd, W.A.; Smith, M.V.; Freedman, J.H. Caenorhabditis elegans as a Model in Developmental Toxicology. Methods Mol. Biol. 2012, 889, 15–24. [Google Scholar] [CrossRef]
- Haynes, W. Bonferroni Correction. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 154. ISBN 978-1-4419-9863-7. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Merritt, C.; Seydoux, G. Transgenic Solutions for the Germline; WormBook: Pasadena, CA, USA, 2010; pp. 1–21. [Google Scholar] [CrossRef]
- Kocsisova, Z.; Kornfeld, K.; Schedl, T. Cell Cycle Accumulation of the Proliferating Cell Nuclear Antigen PCN-1 Transitions from Continuous in the Adult Germline to Intermittent in the Early Embryo of C. elegans. BMC Dev. Biol. 2018, 18, 12. [Google Scholar] [CrossRef]
- Yu, W.; Long, H.; Gao, J.; Wang, Y.; Tu, Y.; Sun, L.; Chen, N. Study on Caenorhabditis elegans as a Combined Model of Microdosimetry and Biology. Dose Response 2021, 19, 1559325821990125. [Google Scholar] [CrossRef]
- Weidhaas, J.B.; Eisenmann, D.M.; Holub, J.M.; Nallur, S.V. A Caenorhabditis elegans Tissue Model of Radiation-Induced Reproductive Cell Death. Proc. Natl. Acad. Sci. USA 2006, 103, 9946–9951. [Google Scholar] [CrossRef]
- Rinaldo, C.; Bazzicalupo, P.; Ederle, S.; Hilliard, M.; La Volpe, A. Roles for Caenorhabditis elegans Rad-51 in Meiosis and in Resistance to Ionizing Radiation during Development. Genetics 2002, 160, 471–479. [Google Scholar] [CrossRef]
- McKay, S.J.; Johnsen, R.; Khattra, J.; Asano, J.; Baillie, D.L.; Chan, S.; Dube, N.; Fang, L.; Goszczynski, B.; Ha, E.; et al. Gene Expression Profiling of Cells, Tissues, and Developmental Stages of the Nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003, 68, 159–169. [Google Scholar] [CrossRef]
- Dupuy, D.; Bertin, N.; Hidalgo, C.A.; Venkatesan, K.; Tu, D.; Lee, D.; Rosenberg, J.; Svrzikapa, N.; Blanc, A.; Carnec, A.; et al. Genome-Scale Analysis of In Vivo Spatiotemporal Promoter Activity in Caenorhabditis elegans. Nat. Biotechnol. 2007, 25, 663–668. [Google Scholar] [CrossRef]
- Shim, J.; Im, S.H.; Lee, J. Tissue-Specific Expression, Heat Inducibility, and Biological Roles of Two Hsp16 Genes in Caenorhabditis elegans. FEBS Lett. 2003, 537, 139–145. [Google Scholar] [CrossRef]
- Candido, E.P.; Jones, D.; Dixon, D.K.; Graham, R.W.; Russnak, R.H.; Kay, R.J. Structure, Organization, and Expression of the 16-kDa Heat Shock Gene Family of Caenorhabditis elegans. Genome 1989, 31, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Stringham, E.G.; Dixon, D.K.; Jones, D.; Candido, E.P. Temporal and Spatial Expression Patterns of the Small Heat Shock (Hsp16) Genes in Transgenic Caenorhabditis elegans. Mol. Biol. Cell 1992, 3, 221–233. [Google Scholar] [CrossRef]
- Jones, D.; Dixon, D.K.; Graham, R.W.; Candido, E.P. Differential Regulation of Closely Related Members of the Hsp16 Gene Family in Caenorhabditis elegans. DNA 1989, 8, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Yanase, S.; Ishi, N. Cloning of the Oxidative Stress-Responsive Genes in Caenorhabditis elegans. J. Radiat. Res. 1999, 40, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Yanase, S.; Hartman, P.S.; Ito, A.; Ishii, N. Oxidative Stress Pretreatment Increases the X-Radiation Resistance of the Nematode Caenorhabditis elegans. Mutat. Res. 1999, 426, 31–39. [Google Scholar] [CrossRef]
- Boyd, W.A.; Crocker, T.L.; Rodriguez, A.M.; Leung, M.C.K.; Wade Lehmann, D.; Freedman, J.H.; Van Houten, B.; Meyer, J.N. Nucleotide Excision Repair Genes Are Expressed at Low Levels and Are Not Detectably Inducible in Caenorhabditis elegans Somatic Tissues, but Their Function Is Required for Normal Adult Life after UVC Exposure. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2010, 683, 57–67. [Google Scholar] [CrossRef]
- Pujol, N.; Cypowyj, S.; Ziegler, K.; Millet, A.; Astrain, A.; Goncharov, A.; Jin, Y.; Chisholm, A.D.; Ewbank, J.J. Distinct Innate Immune Responses to Infection and Wounding in the C. elegans Epidermis. Curr. Biol. 2008, 18, 481–489. [Google Scholar] [CrossRef]
- Hegedus, F.; Mathew, L.M.; Schwartz, R.A. Radiation Dermatitis: An Overview. Int. J. Dermatol. 2017, 56, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Panizzon, R. Radiodermatitis: State of the Art. J. Eur. Acad. Dermatol. Venereol. 2004, 18, 12. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beaudier, P.; Devès, G.; Plawinski, L.; Dupuy, D.; Barberet, P.; Seznec, H. Proton Microbeam Targeted Irradiation of the Gonad Primordium Region Induces Developmental Alterations Associated with Heat Shock Responses and Cuticle Defense in Caenorhabditis elegans. Biology 2023, 12, 1372. https://doi.org/10.3390/biology12111372
Beaudier P, Devès G, Plawinski L, Dupuy D, Barberet P, Seznec H. Proton Microbeam Targeted Irradiation of the Gonad Primordium Region Induces Developmental Alterations Associated with Heat Shock Responses and Cuticle Defense in Caenorhabditis elegans. Biology. 2023; 12(11):1372. https://doi.org/10.3390/biology12111372
Chicago/Turabian StyleBeaudier, Pierre, Guillaume Devès, Laurent Plawinski, Denis Dupuy, Philippe Barberet, and Hervé Seznec. 2023. "Proton Microbeam Targeted Irradiation of the Gonad Primordium Region Induces Developmental Alterations Associated with Heat Shock Responses and Cuticle Defense in Caenorhabditis elegans" Biology 12, no. 11: 1372. https://doi.org/10.3390/biology12111372
APA StyleBeaudier, P., Devès, G., Plawinski, L., Dupuy, D., Barberet, P., & Seznec, H. (2023). Proton Microbeam Targeted Irradiation of the Gonad Primordium Region Induces Developmental Alterations Associated with Heat Shock Responses and Cuticle Defense in Caenorhabditis elegans. Biology, 12(11), 1372. https://doi.org/10.3390/biology12111372