Enhanced Herbicide Metabolism and Target-Site Mutations Confer Multiple Resistance to Fomesafen and Nicosulfuron in Amaranthus retroflexus L.
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Whole-Plant Dose–Response Experiments
2.3. Gene Sequencing and In Vitro Assay of ALS Activity
2.4. Effect of P450 and GST Inhibitors on Fomesafen and Nicosulfuron Resistance
2.5. Analysis of Fomesafen and Nicosulfuron Metabolism in A. retroflexus
2.6. Statistical Analyses
3. Results
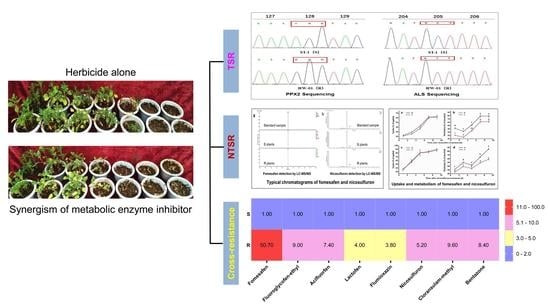
3.1. Multiple Resistance to Fomesafen and Nicosulfuron and Target-Site Mutation(s) Identification in A. retroflexus
3.2. In Vitro Assay of ALS for Nicosulfuron Activity
3.3. Impact of P450 and GST Inhibitors on Fomesafen and Nicosulfuron Resistance
3.4. Fomesafen and Nicosulfuron Metabolism in A. retroflexus
3.5. Dose Response to Other Herbicides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Guo, W.; Zhang, L.; Zhao, K.; Ge, L.; Lv, X.; Liu, W.; Wang, J. Multiple resistance to thifensulfuron-methyl and fomesafen in redroot pigweed (Amaranthus retroflexus L.) from China. Chil. J. Agric. Res. 2017, 77, 311–317. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Zhao, N.; Zhu, B.; Sun, P.; Liu, W.; Wang, J. Multiple resistance to PPO and ALS inhibitors in redroot pigweed (Amaranthus retroflexus). Weed Sci. 2020, 68, 19–26. [Google Scholar] [CrossRef]
- Baskin, C.C. Germination ecology of seeds in the persistant seed bank. In Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination; Baskin, C.C., Baskin, J.M., Eds.; Academic Press: San Diego, CA, USA, 1998; pp. 133–179. [Google Scholar]
- Bensch, C.N.; Horak, M.J.; Peterson, D. Interference of redroot pigweed (Amaranthus retroflexus), Palmer amaranth (A. palmeri), and common waterhemp (A. rudis) in soybean. Weed Sci. 2003, 51, 37–43. [Google Scholar] [CrossRef]
- Ghanizadeh, H.; Lorzadeh, S.; Aryannia, N. Effect of weed interference on Zea mays: Growth analysis. Weed Biol. Manag. 2014, 14, 133–137. [Google Scholar] [CrossRef]
- Sheibany, K.; Baghestani Meybodi, M.A.; Atri, A. Competitive effects of redroot pigweed (Amaranthus retroflexus) on the growth indices and yield of corn. Weed Biol. Manag. 2009, 9, 152–159. [Google Scholar] [CrossRef]
- Aguyoh, J.N.; Masiunas, J.B. Interference of redroot pigweed (Amaranthus retroflexus) with snap beans. Weed Sci. 2003, 51, 202–207. [Google Scholar] [CrossRef]
- Beale, S.I. Tetrapyrrole metabolism in photosynthetic organisms. In Biosynthesis of Heme and Chlorophylls; Dailey, H.A., Ed.; McGraw-Hill: New York, NY, USA, 1990. [Google Scholar]
- Jacobs, J.M.; Jacobs, N.J. Porphyrin Accumulation and Export by Isolated Barley (Hordeum vulgare) Plastids (Effect of Diphenyl Ether Herbicides). Plant Physiol. 1993, 101, 1181–1187. [Google Scholar] [CrossRef]
- Duke, S.O.; Lydon, J.; Becerril, J.M.; Sherman, T.D.; Lehnen, L.P.; Matsumoto, H. Protoporphyrinogen Oxidase-Inhibiting Herbicides. Weed Sci. 1991, 39, 465–473. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, S.B.; Chung, J.S.; Han, S.U.; Han, O.; Guh, J.O.; Jeon, J.S.; An, G.; Back, K. Transgenic Rice Plants Expressing a Bacillus subtilis Protoporphyrinogen Oxidase Gene Are Resistant to Diphenyl Ether Herbicide Oxyfluorfen. Plant Cell Physiol. 2000, 41, 743–749. [Google Scholar] [CrossRef]
- Mei, Y.; Si, C.; Liu, M.; Qiu, L.; Zheng, M. Investigation of resistance levels and mechanisms to nicosulfuron conferred by non-target-site mechanisms in large crabgrass (Digitaria sanguinalis L.) from China. Pestic. Biochem. Physiol. 2017, 141, 84–89. [Google Scholar] [CrossRef]
- Heap, I. The International Survey of Herbicide Resistant Weeds. Available online: www.weedscience.org (accessed on 4 April 2023).
- Patzoldt, W.L.; Hager, A.G.; McCormick, J.S.; Tranel, P.J. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc. Natl. Acad. Sci. USA 2006, 103, 12329–12334. [Google Scholar] [CrossRef]
- Salas, R.A.; Burgos, N.R.; Tranel, P.J.; Singh, S.; Glasgow, L.; Scott, R.C.; Nichols, R.L. Resistance to PPO-inhibiting herbicide in Palmer amaranth from Arkansas. Pest Manag. Sci. 2016, 72, 864–869. [Google Scholar] [CrossRef]
- Bi, B.; Wang, Q.; Coleman, J.J.; Porri, A.; Peppers, J.M.; Patel, J.D.; Betz, M.; Lerchl, J.; McElroy, J.S. A novel mutation A212T in chloroplast Protoporphyrinogen oxidase (PPO1) confers resistance to PPO inhibitor Oxadiazon in Eleusine indica. Pest Manag. Sci. 2020, 76, 1786–1794. [Google Scholar] [CrossRef]
- Giacomini, D.A.; Umphres, A.M.; Nie, H.; Mueller, T.C.; Steckel, L.E.; Young, B.G.; Scott, R.C.; Tranel, P.J. Two new PPX2 mutations associated with resistance to PPO-inhibiting herbicides in Amaranthus palmeri. Pest Manag. Sci. 2017, 73, 1559–1563. [Google Scholar] [CrossRef]
- Mendes, R.R.; Takano, H.K.; Adegas, F.S.; Oliveira, R.S.; Gaines, T.A.; Dayan, F.E. Arg-128-Leu target-site mutation in PPO2 evolves in wild poinsettia (Euphorbia heterophylla) with cross-resistance to PPO-inhibiting herbicides. Weed Sci. 2020, 68, 437–444. [Google Scholar] [CrossRef]
- Rangani, G.; Salas-Perez, R.A.; Aponte, R.A.; Knapp, M.; Craig, I.R.; Mietzner, T.; Langaro, A.C.; Noguera, M.M.; Porri, A.; Roma-Burgos, N. A Novel Single-Site Mutation in the Catalytic Domain of Protoporphyrinogen Oxidase IX (PPO) Confers Resistance to PPO-Inhibiting Herbicides. Front. Plant Sci. 2019, 10, 568. [Google Scholar] [CrossRef]
- Rousonelos, S.L.; Lee, R.M.; Moreira, M.S.; VanGessel, M.J.; Tranel, P.J. Characterization of a Common Ragweed (Ambrosia artemisiifolia) Population Resistant to ALS- and PPO-Inhibiting Herbicides. Weed Sci. 2012, 60, 335–344. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R.; Heap, I. Mutations in Herbicide-Resistant Weeds to Inhibition of Acetolactate Synthase. Available online: http://www.weedscience.com (accessed on 4 April 2023).
- Chen, J.; Huang, Z.; Zhang, C.; Huang, H.; Wei, S.; Chen, J.; Wang, X. Molecular basis of resistance to imazethapyr in redroot pigweed (Amaranthus retroflexus L.) populations from China. Pestic. Biochem. Physiol. 2015, 124, 43–47. [Google Scholar] [CrossRef]
- Du, L.; Li, X.; Jiang, X.; Ju, Q.; Guo, W.; Li, L.; Qu, C.; Qu, M. Target-site basis for fomesafen resistance in redroot pigweed (Amaranthus retroflexus) from China. Weed Sci. 2021, 69, 290–299. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-Based Herbicide Resistance and Cross-Resistance in Crop Weeds: A Threat to Herbicide Sustainability and Global Crop Production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Ghanizadeh, H.; Harrington, K.C. Non-target Site Mechanisms of Resistance to Herbicides. Crit. Rev. Plant Sci. 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Varanasi, V.K.; Brabham, C.; Norsworthy, J.K. Confirmation and Characterization of Non–target site Resistance to Fomesafen in Palmer amaranth (Amaranthus palmeri). Weed Sci. 2018, 66, 702–709. [Google Scholar] [CrossRef]
- Han, H.; Yu, Q.; Purba, E.; Li, M.; Walsh, M.; Friesen, S.; Powles, S.B. A novel amino acid substitution Ala-122-Tyr in ALS confers high-level and broad resistance across ALS-inhibiting herbicides. Pest Manag. Sci. 2012, 68, 1164–1170. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Vila-Aiub, M.M.; Powles, S.B. AHAS herbicide resistance endowing mutations: Effect on AHAS functionality and plant growth. J. Exp. Bot. 2010, 61, 3925–3934. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Gaines, T.A.; Dayan, F.E.; Patterson, E.L.; Jhala, A.J.; Knezevic, S.Z. Reversing resistance to tembotrione in an Amaranthus tuberculatus (var. rudis) population from Nebraska, USA with cytochrome P450 inhibitors. Pest Manag. Sci. 2018, 74, 2296–2305. [Google Scholar] [CrossRef]
- Ma, R.; Evans, A.F.; Riechers, D.E. Differential Responses to Preemergence and Postemergence Atrazine in Two Atrazine-Resistant Waterhemp Populations. Agron. J. 2016, 108, 1196–1202. [Google Scholar] [CrossRef]
- Ma, R.; Kaundun, S.S.; Tranel, P.J.; Riggins, C.W.; McGinness, D.L.; Hager, A.G.; Hawkes, T.; McIndoe, E.; Riechers, D.E. Distinct Detoxification Mechanisms Confer Resistance to Mesotrione and Atrazine in a Population of Waterhemp. Plant Physiol. 2013, 163, 363–377. [Google Scholar] [CrossRef]
- Bai, S.; Liu, W.; Wang, H.; Zhao, N.; Jia, S.; Zou, N.; Guo, W.; Wang, J. Enhanced Herbicide Metabolism and Metabolic Resistance Genes Identified in Tribenuron-Methyl Resistant Myosoton aquaticum L. J. Agric. Food Chem. 2018, 66, 9850–9857. [Google Scholar] [CrossRef]
- Zhao, J. Production and field application status of Diphenylethers herbicides. Chin. J. Pestic. 2019, 237–238. [Google Scholar] [CrossRef]
- Dayan, F.E.; Barker, A.; Tranel, P.J. Origins and structure of chloroplastic and mitochondrial plant protoporphyrinogen oxidases: Implications for the evolution of herbicide resistance. Pest Manag. Sci. 2018, 74, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Lamego, F.P.; Charlson, D.; Delatorre, C.A.; Burgos, N.R.; Vidal, R.A. Molecular Basis of Resistance to ALS-Inhibitor Herbicides in Greater Beggarticks. Weed Sci. 2009, 57, 474–481. [Google Scholar] [CrossRef]
- Jasieniuk, M.; Brûlé-Babel, A.L.; Morrison, I.N. The Evolution and Genetics of Herbicide Resistance in Weeds. Weed Sci. 1996, 44, 176–193. [Google Scholar] [CrossRef]
- Preston, C. Herbicide resistance in weeds endowed by enhanced detoxification: Complications for management. Weed Sci. 2004, 52, 448–453. [Google Scholar] [CrossRef]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef]
- Duke, S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 2012, 68, 505–512. [Google Scholar] [CrossRef]
- Busi, R.; Gaines, T.A.; Powles, S. Phorate can reverse P450 metabolism-based herbicide resistance in Lolium rigidum. Pest Manag. Sci. 2017, 73, 410–417. [Google Scholar] [CrossRef]
- Preston, C.; Tardif, F.J.; Christopher, J.T.; Powles, S.B. Multiple Resistance to Dissimilar Herbicide Chemistries in a Biotype of Lolium rigidum Due to Enhanced Activity of Several Herbicide Degrading Enzymes. Pestic. Biochem. Physiol. 1996, 54, 123–134. [Google Scholar] [CrossRef]
- Lee, R.M.; Hager, A.G.; Tranel, P.J. Prevalence of a novel resistance mechanism to PPO-inhibiting herbicides in waterhemp (Amaranthus tuberculatus). Weed Sci. 2008, 56, 371–375. [Google Scholar] [CrossRef]
- Rangani, G.; Noguera, M.; Salas-Perez, R.; Benedetti, L.; Roma-Burgos, N. Mechanism of resistance to S-metolachlor in Palmer amaranth. Front. Plant Sci. 2021, 12, 652581. [Google Scholar] [CrossRef] [PubMed]
- Küpper, A.; Peter, F.; Zöllner, P.; Lorentz, L.; Tranel, P.J.; Beffa, R.; Gaines, T.A. Tembotrione detoxification in 4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor-resistant Palmer amaranth (Amaranthus palmeri S. Wats.). Pest Manag. Sci. 2018, 74, 2325–2334. [Google Scholar] [CrossRef]
- Obenland, O.A.; Ma, R.; O’Brien, S.R.; Lygin, A.V.; Riechers, D.E. Carfentrazone-ethyl resistance in an Amaranthus tuberculatus population is not mediated by amino acid alterations in the PPO2 protein. PLoS ONE 2019, 14, e0215431. [Google Scholar] [CrossRef]
- Huang, Z.; Cui, H.; Wang, C.; Wu, T.; Zhang, C.; Huang, H.; Wei, S. Investigation of resistance mechanism to fomesafen in Amaranthus retroflexus L. Pestic. Biochem. Physiol. 2020, 165, 104560. [Google Scholar] [CrossRef]
- Bi, Y.; Liu, W.; Guo, W.; Li, L.; Yuan, G.; Du, L.; Wang, J. Molecular basis of multiple resistance to ACCase- and ALS-inhibiting herbicides in Alopecurus japonicus from China. Pestic. Biochem. Physiol. 2016, 126, 22–27. [Google Scholar] [CrossRef]
- Fang, J.; Zhang, Y.; Liu, T.; Yan, B.; Li, J.; Dong, L. Target-Site and Metabolic Resistance Mechanisms to Penoxsulam in Barnyardgrass (Echinochloa crus-galli (L.) P. Beauv). J. Agric. Food Chem. 2019, 67, 8085–8095. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Ge, L.a.; Zhu, B.; Liu, W.; Wang, J. Target site mutations and cytochrome P450s confer resistance to fenoxaprop-P-ethyl and mesosulfuron-methyl in Alopecurus aequalis. Pest Manag. Sci. 2019, 75, 204–214. [Google Scholar] [CrossRef]
- Tan, J.; Li, Q.; Guo, W.; Liu, W.; Wang, J. Evaluation of herbicidal activity and safety to cotton of flumioxazin. Chin. J. Pestic. Sci. 2017, 19, 189–194. [Google Scholar] [CrossRef]
- Owen, M.J.; Martinez, N.J.; Powles, S.B. Multiple herbicide-resistant wild radish (Raphanus raphanistrum) populations dominate Western Australian cropping fields. Crop Pasture Sci. 2015, 66, 1079–1085. [Google Scholar] [CrossRef]
- Walsh, M.J.; Powles, S.B.; Beard, B.R.; Parkin, B.T.; Porter, S.A. Multiple-herbicide resistance across four modes of action in wild radish (Raphanus raphanistrum). Weed Sci. 2004, 52, 8–13. [Google Scholar] [CrossRef]
- Scarabel, L.; Varotto, S.; Sattin, M. A European biotype of Amaranthus retroflexus cross-resistant to ALS inhibitors and response to alternative herbicides. Weed Res. 2007, 47, 527–533. [Google Scholar] [CrossRef]





| Herbicide a | Group b | Application Rate (g ai ha−1) | |
|---|---|---|---|
| ST-1 (S) | HW-01 (R) | ||
| Fomesafen | E | 0.36, 1.8, 9, 45, 225, 1125 | 3, 15, 75, 375, 1875, 9375 |
| Fluoroglycofen-ethyl | E | 0.096, 0.48, 2.4, 12, 60, 300 | 0.72, 3.6, 18, 90, 450, 2250 |
| Acifluorfen | E | 0.58, 2.89, 14.4, 71.9, 359.5, 1797.6 | 3.9, 19.3, 96.3, 481.5, 2407.5, 12,037.5 |
| Lactofen | E | 0.72, 3.6, 18, 90, 450, 2250 | 1.01, 5.04, 25.2, 126, 630, 3150 |
| Flumioxazin | E | 0.18, 0.9, 4.5, 22.5, 112.5, 562.5 | 0.24, 1.2, 6, 30, 150, 750 |
| Bentazone | C3 | 5.9, 29.9, 149.6, 748, 3740, 18,700 | 11.9, 59.9, 299.5, 1497.6, 7488, 37,440 |
| Nicosulfuron | B | 0.384, 1.92, 9.6, 48, 240, 1200 | 0.48, 2.4, 12, 60, 300, 1500 |
| Cloransulam-methyl | B | 0.04, 0.20, 1.0, 5.0, 25.0, 125.0 | 0.2, 1.0, 5.0, 25.2, 126, 630 |
| Herbicide | Retention Time (min) | Quantitative Ions | Qualitative Ion | Collision Energy (eV) | Scan Mode |
|---|---|---|---|---|---|
| Fomesafen | 2.52 | 437.05/194.50 | 437.05/222.00 | 36.26 a/31.45 | ESI− |
| Nicosulfuron | 7.76 | 411.06/182.17 | 411.06/213.18 | 17 a/15 | ESI+ |
| Herbicide | Group a | Populations b | Regression Parameters c | GR50 | RId | |||
|---|---|---|---|---|---|---|---|---|
| c | d | b | r2 | |||||
| Fomesafen | E | R | 0.18 (0.005) | 93.13 (0.004) | −1.10 (0.12) | 0.9989 | 235.71 (24.46) | 50.7 |
| S | 7.37 (1.83) | 98.90 (3.21) | −2.89 (0.55) | 0.9980 | 4.65 (0.67) | |||
| Fluoroglycofen-ethyl | E | R | 4.69 (1.56) | 96.08 (5.52) | −0.75 (0.20) | 0.9949 | 90.84 (3.90) | 9.0 |
| S | 6.47 (1.07) | 98.66 (4.36) | −0.83 (0.16) | 0.9970 | 10.10 (2.27) | |||
| Acifluorfen | E | R | 6.15 (2.59) | 98.40 (6.27) | −0.81 (0.23) | 0.9936 | 447.67 (53.39) | 7.4 |
| S | 7.53 (1.96) | 97.47 (3.96) | −0.81 (0.42) | 0.9975 | 60.39 (13.09) | |||
| Lactofen | E | R | 9.66 (0.26) | 93.87 (0.29) | −1.28 (0.02) | 1.0000 | 42.61 (0.59) | 4.0 |
| S | 4.32 (1.46) | 96.03 (2.74) | −1.62 (0.19) | 0.9988 | 10.78 (0.99) | |||
| Flumioxazin | E | R | 9.99 (1.50) | 96.15 (2.62) | −3.13 (0.54) | 0.9985 | 2.28 (0.29) | 3.8 |
| S | 16.28 (2.41) | 84.09 (4.19) | −2.84 (1.77) | 0.9934 | 0.60 (0.17) | |||
| Nicosulfuron | B | R | 13.52 (3.43) | 94.46 (4.18) | −0.90 (0.17) | 0.9922 | 19.21 (3.91) | 5.2 |
| S | 11.51 (2.32) | 96.38 (4.91) | −1.57 (0.29) | 0.9896 | 3.69 (0.61) | |||
| Cloransulam-methyl | B | R | 1.12 (0.16) | 95.23 (1.52) | −1.00 (0.09) | 0.9992 | 30.57 (2.96) | 9.6 |
| S | 0.83 (2.56) | 96.41 (2.07) | −0.87 (0.08) | 0.9992 | 3.20 (0.33) | |||
| Bentazone | C3 | R | 9.40 (3.72) | 101.49 (5.33) | −0.83 (0.15) | 0.9974 | 381.26 (78.48) | 8.4 |
| S | 11.22 (3.75) | 112.93 (13.97) | −0.96 (0.27) | 0.9936 | 45.52 (17.65) | |||
| Herbicide | HW-01 | ST-1 | ||||
|---|---|---|---|---|---|---|
| GR50 | GR50 Reduction (%) | p-Value | GR50 | GR50 Reduction (%) | p-Value | |
| Fomesafen | 235.71 (24.46) | -- | -- | 4.65 (0.67) | -- | -- |
| Fomesafen + Malathion | 40.51 (6.15) * | 82.83% | 0.00 | 4.58 (0.46) | 1.51% | 0.99 |
| Fomesafen +PBO | 47.91 (3.20) * | 79.67% | 0.00 | 4.39 (0.76) | 5.59% | 0.81 |
| Fomesafen + Amitrole | 71.98 (13.08) * | 69.46% | 0.00 | 4.41 (0.54) | 5.16% | 0.82 |
| Fomesafen + NBD-Cl | 85.12 (19.54) * | 63.89% | 0.00 | 4.49 (0.53) | 3.44% | 0.93 |
| Nicosulfuron | 19.21 (3.91) | -- | -- | 3.69 (0.61) | -- | |
| Nicosulfuron + Malathion | 6.61 (1.42) * | 65.59% | 0.00 | 3.37 (0.58) | 8.67% | 0.65 |
| Nicosulfuron + PBO | 9.04 (2.00) * | 52.94% | 0.00 | 3.57 (0.42) | 3.25% | 0.96 |
| Nicosulfuron + Amitrole | 7.76 (2.38) * | 59.60% | 0.00 | 3.56 (0.58) | 3.52% | 0.95 |
| Nicosulfuron + NBD-Cl | 6.73 (1.68) * | 64.97% | 0.00 | 3.49 (0.33) | 5.42% | 0.86 |
| DAT a | Residual Dose of Fomesafen (μg) | Uptake Dose of Fomesafen (μg) | Metabolism Dose of Fomesafen (μg) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HW-01 | ST-1 | p-Value | HW-01 | ST-1 | p-Value | HW-01 | ST-1 | p-Value | |
| 1 | 2.60 (1.40) | 3.39 (0.11) | 0.22 | 4.47 (1.11) | 4.26 (1.27) | 0.94 | 1.88 (0.23) | 0.88 (0.14) * | 0.00 |
| 3 | 3.96 (1.22) | 4.34 (1.13) | 0.75 | 5.89 (1.23) | 5.07 (1.15) | 0.17 | 1.94 (0.47) | 0.73 (0.17) * | 0.00 |
| 5 | 4.08 (1.63) | 4.95 (1.29) | 0.23 | 8.48 (1.49) | 7.54 (1.94) | 0.28 | 4.40 (1.44) | 2.59 (0.40) * | 0.00 |
| 7 | 2.67 (1.08) | 4.61 (1.53) * | 0.00 | 12.41 (1.98) | 11.96 (2.21) | 0.87 | 9.74 (1.87) | 7.35 (1.00) * | 0.00 |
| 9 | 2.75 (1.07) | 4.65 (1.54) * | 0.00 | 12.56 (2.05) | 12.33 (3.54) | 0.99 | 9.81 (1.60) | 7.68 (1.28) * | 0.00 |
| DAT a | Residual Dose of Nicosulfuron (μg) | Uptake Dose of Nicosulfuron (μg) | Metabolism Dose of Nicosulfuron (μg) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HW-01 | ST-1 | p-Value | HW-01 | ST-1 | p-Value | HW-01 | ST-1 | p-Value | |
| 1 | 1.20 (0.23) | 1.42 (0.37) | 0.73 | 1.75 (0.32) | 1.87 (0.08) | 0.42 | 0.55 (0.04) | 0.45 (0.06) * | 0.01 |
| 3 | 1.46 (0.27) | 1.68 (0.18) | 0.14 | 2.92 (0.41) | 2.76 (0.13) | 0.40 | 1.46 (0.21) | 1.08 (0.11) * | 0.00 |
| 5 | 2.32 (0.53) | 2.64 (0.42) | 0.30 | 4.02 (0.86) | 3.99 (0.26) | 0.93 | 1.70 (0.36) | 1.35 (0.17) | 0.07 |
| 7 | 1.49 (0.16) | 2.11 (0.08) * | 0.00 | 4.19 (0.22) | 4.12 (0.24) | 0.63 | 2.70 (0.09) | 2.01 (0.09) * | 0.00 |
| 9 | 0.62 (0.09) | 1.83 (0.15) * | 0.00 | 4.48 (0.31) | 4.41 (0.35) | 0.73 | 3.86 (0.12) | 2.58 (0.20) * | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Wang, H.; Duan, Y.; Bei, F.; Jia, S.; Wang, J.; Wang, H.; Liu, W. Enhanced Herbicide Metabolism and Target-Site Mutations Confer Multiple Resistance to Fomesafen and Nicosulfuron in Amaranthus retroflexus L. Biology 2023, 12, 592. https://doi.org/10.3390/biology12040592
Yang C, Wang H, Duan Y, Bei F, Jia S, Wang J, Wang H, Liu W. Enhanced Herbicide Metabolism and Target-Site Mutations Confer Multiple Resistance to Fomesafen and Nicosulfuron in Amaranthus retroflexus L. Biology. 2023; 12(4):592. https://doi.org/10.3390/biology12040592
Chicago/Turabian StyleYang, Cheng, Hao Wang, Yunxia Duan, Feng Bei, Sisi Jia, Jinxin Wang, Hengzhi Wang, and Weitang Liu. 2023. "Enhanced Herbicide Metabolism and Target-Site Mutations Confer Multiple Resistance to Fomesafen and Nicosulfuron in Amaranthus retroflexus L." Biology 12, no. 4: 592. https://doi.org/10.3390/biology12040592
APA StyleYang, C., Wang, H., Duan, Y., Bei, F., Jia, S., Wang, J., Wang, H., & Liu, W. (2023). Enhanced Herbicide Metabolism and Target-Site Mutations Confer Multiple Resistance to Fomesafen and Nicosulfuron in Amaranthus retroflexus L. Biology, 12(4), 592. https://doi.org/10.3390/biology12040592