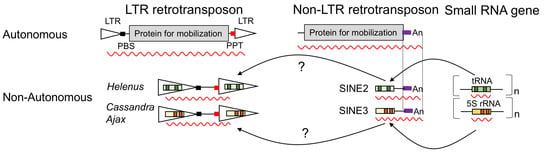
Helenus and Ajax, Two Groups of Non-Autonomous LTR Retrotransposons, Represent a New Type of Small RNA Gene-Derived Mobile Elements
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening of TRIMs with tRNA-Derived Sequences
2.2. Prediction of tRNA Primer Usage and Similarity
2.3. Screening of TRIMs with 5S rRNA-Derived Sequences
2.4. Characterization of SINE3 from Lepidopteran Genomes
2.5. Estimation of Copy Numbers
3. Results
3.1. Helenus: TRIM with a tRNA-Derived Sequence
3.2. Helenus from Fungi
3.3. Helenus from Animals
3.4. Helenus from Plants
3.5. tRNA-Derived Sequences in Helenus
3.6. Primer-Binding Sites (PBSs) of Helenus
3.7. Ajax: TRIM with a 5S rRNA Promoter
3.8. Copy Number Estimates of Helenus and Ajax
4. Discussion
4.1. TRIMp3—Proposed TRIM Families Containing an RNA Polymerase III Promoter
4.2. The Evolution of TRIMp3
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2020, 94, 233–252. [Google Scholar] [CrossRef]
- Arkhipova, I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 2017, 8, 19. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Luo, X.; Chen, S.; Zhang, Y. PlantRep: A database of plant repetitive elements. Plant Cell Rep. 2022, 41, 1163–1166. [Google Scholar] [CrossRef]
- Stitzer, M.C.; Anderson, S.N.; Springer, N.M.; Ross-Ibarra, J. The genomic ecosystem of transposable elements in maize. PLoS Genet. 2021, 17, e1009768. [Google Scholar] [CrossRef]
- Curcio, M.J.; Derbyshire, K.M. The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003, 4, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Telesnitsky, A.; Goff, S.P. Reverse Transcriptase and the Generation of Retroviral DNA. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Kajikawa, M.; Okada, N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 2002, 111, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Vassetzky, N.S.; Kramerov, D.A. SINEBase: A database and tool for SINE analysis. Nucleic Acids Res. 2013, 41, D83–D89. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Okada, N. Generality of the tRNA origin of short interspersed repetitive elements (SINEs). Characterization of three different tRNA-derived retroposons in the octopus. J. Mol. Biol. 1994, 243, 25–37. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L.; Hellmann-Blumberg, U.; Jurka, J.; Labuda, D.; Rubin, C.M.; Schmid, C.W.; Zietkiewicz, E.; Zuckerkandl, E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996, 42, 3–6. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. A novel class of SINE elements derived from 5S rRNA. Mol. Biol. Evol. 2003, 20, 694–702. [Google Scholar] [CrossRef]
- Kojima, K.K. Hagfish genome reveals parallel evolution of 7SL RNA-derived SINEs. Mob. DNA 2020, 11, 18. [Google Scholar] [CrossRef]
- Wang, J.; Wang, A.; Han, Z.; Zhang, Z.; Li, F.; Li, X. Characterization of three novel SINE families with unusual features in Helicoverpa armigera. PLoS ONE 2012, 7, e31355. [Google Scholar] [CrossRef]
- Witte, C.P.; Le, Q.H.; Bureau, T.; Kumar, A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 2001, 98, 13778–13783. [Google Scholar] [CrossRef]
- Gao, D.; Chen, J.; Chen, M.; Meyers, B.C.; Jackson, S. A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PLoS ONE 2012, 7, e32010. [Google Scholar] [CrossRef]
- Gao, D.; Li, Y.; Kim, K.D.; Abernathy, B.; Jackson, S.A. Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol. 2016, 17, 7. [Google Scholar] [CrossRef]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; de Graaf, D.C.; Dearden, P.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.; Hasselmann, M.; Lozier, J.D.; et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef]
- Zhou, Y.; Cahan, S.H. A novel family of terminal-repeat retrotransposon in miniature (TRIM) in the genome of the red harvester ant, Pogonomyrmex barbatus. PLoS ONE 2012, 7, e53401. [Google Scholar] [CrossRef]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef]
- Koziol, U.; Radio, S.; Smircich, P.; Zarowiecki, M.; Fernandez, C.; Brehm, K. A Novel Terminal-Repeat Retrotransposon in Miniature (TRIM) Is Massively Expressed in Echinococcus multilocularis Stem Cells. Genome Biol. Evol. 2015, 7, 2136–2153. [Google Scholar] [CrossRef]
- Satovic, E.; Luchetti, A.; Pasantes, J.J.; Garcia-Souto, D.; Cedilak, A.; Mantovani, B.; Plohl, M. Terminal-Repeat Retrotransposons in Miniature (TRIMs) in bivalves. Sci. Rep. 2019, 9, 19962. [Google Scholar] [CrossRef]
- Kalendar, R.; Tanskanen, J.; Chang, W.; Antonius, K.; Sela, H.; Peleg, O.; Schulman, A.H. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 5833–5838. [Google Scholar] [CrossRef]
- Kalendar, R.; Raskina, O.; Belyayev, A.; Schulman, A.H. Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants. Int. J. Mol. Sci. 2020, 21, 2931. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Aime, M.C.; Bell, C.D.; Wilson, A.W. Deconstructing the evolutionary complexity between rust fungi (Pucciniales) and their plant hosts. Stud. Mycol. 2018, 89, 143–152. [Google Scholar] [CrossRef]
- Cruaud, A.; Rasplus, J.Y.; Zhang, J.; Burks, R.; Delvare, G.; Fusu, L.; Gumovsky, A.; Huber, J.T.; Jansta, P.; Mitroiu, M.D.; et al. The Chalcidoidea bush of life: Evolutionary history of a massive radiation of minute wasps. Cladistics 2023, 40, 34–63. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, M.; Noguchi, H.; Nishihara, H.; Toyoda, A.; Suzuki, Y.; Kajitani, R.; Suzuki, H.; Okuno, M.; Aibara, M.; Ngatunga, B.P.; et al. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013, 23, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef]
- Combosch, D.J.; Giribet, G. Clarifying phylogenetic relationships and the evolutionary history of the bivalve order Arcida (Mollusca: Bivalvia: Pteriomorphia). Mol. Phylogenet Evol. 2016, 94, 298–312. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Ando, Y.; Shiba, T. Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. Nature 1986, 323, 824–826. [Google Scholar] [CrossRef]
- Ke, N.; Gao, X.; Keeney, J.B.; Boeke, J.D.; Voytas, D.F. The yeast retrotransposon Ty5 uses the anticodon stem-loop of the initiator methionine tRNA as a primer for reverse transcription. RNA 1999, 5, 929–938. [Google Scholar] [CrossRef]
- Neumann, P.; Novak, P.; Hostakova, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Han, G.; Zhang, N.; Jiang, H.; Meng, X.; Qian, K.; Zheng, Y.; Xu, J.; Wang, J. Diversity of short interspersed nuclear elements (SINEs) in lepidopteran insects and evidence of horizontal SINE transfer between baculovirus and lepidopteran hosts. BMC Genom. 2021, 22, 226. [Google Scholar] [CrossRef]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef]
- de Souza, F.S.; Franchini, L.F.; Rubinstein, M. Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol. Biol. Evol. 2013, 30, 1239–1251. [Google Scholar] [CrossRef]
- Zhang, X.O.; Pratt, H.; Weng, Z. Investigating the Potential Roles of SINEs in the Human Genome. Annu. Rev. Genom. Hum. Genet. 2021, 22, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; Urbina, D.; de la Pena, M. Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol. 2016, 17, 135. [Google Scholar] [CrossRef]





| Kingdom | Phylum | Class | Order | Family | Species |
|---|---|---|---|---|---|
| Fungi | Basidiomycota | Pucciniomycetes | Pucciniales | Pucciniaceae | Puccinia graminis, P. striiformis, P. triticina, P. hordei, P. horiana, P. coronata, P. novopanici, P. sorghi, P. arachidis, P. polysora, Uromyces transversalis, and U. viciae-fabae |
| Sphaerophragmiaceae | Austropuccinia psidii | ||||
| Melampsoraceae | Melampsora larici-populina, M. aecidioides, and M. medusae | ||||
| Coleosporiaceae | Cronartium ribicola | ||||
| Zaghouaniaceae | Hemileia vastatrix | ||||
| Agaricomycetes | Agaricales | Agaricaceae | Agaricus bisporus | ||
| Psathyrellaceae | Coprinopsis cinerea | ||||
| Metazoa | Ctenophora | Tentaculata | Lobata | Bolinopsidae | Mnemiopsis leidyi |
| Chordata | - | Coelacanthiformes | Coelacanthidae | Latimeria chalumnae | |
| Actinopteri | Acipenseriformes | Acipenseridae | Acipenser ruthenus | ||
| Polyodontidae | Polyodon spathula | ||||
| Cypriniformes | Danionidae | Danio rerio | |||
| Cyprinidae | Cyprinus carpio, Carassius auratus, Culter alburnus, Onychostoma macrolepis, Phoxinus phoxinus, and Squalius cephalus | ||||
| Nemacheilidae | Triplophysa dalaica | ||||
| Siluriformes | Bagridae | Hemibagrus wyckioides | |||
| Gonorynchiformes | Chanidae | Chanos chanos | |||
| Mollusca | Bivalvia | Ostreida | Ostreidae | Crassostrea gigas, C. virginica, and Saccostrea glomerata | |
| Pterioida | Pinnidae | Pinna nobilis | |||
| Pteriidae | Pinctada imbricata | ||||
| Mytilida | Mytilidae | Mytilus galloprovincialis, M. coruscus, Modiolus philippinarum, Gigantidas (Bathymodiolus) platifrons, and Limnoperna fortunei | |||
| Pectinida | Pectinidae | Argopecten irradians, Mizuhopecten yessoensis, and Pecten maximus | |||
| Arcoida | Arcidae | Tegillarca granosa | |||
| Brachiopoda | Lingulata | Lingulida | Lingulidae | Lingula anatina | |
| Nemertea | Pilidiophora | Heteronemertea | Lineidae | Lineus longissimus and Notospermus geniculatus | |
| Arthropoda | Insecta | Hymenoptera | Pteromalidae | Nasonia vitripennis, Cecidostiba fungosa, Trichomalopsis sarcophagae, Muscidifurax raptorellus, Philotrypesis tridentata, Theocolax elegans, Gastracanthus pulcherrimus, and Pteromalus puparum | |
| Eurytomidae | Eurytoma adleriae | ||||
| Eupelmidae | Eupelmus annulatus | ||||
| Megastigmidae | Megastigmus dorsalis | ||||
| Cynipidae | Synergus japonicus and S. umbraculus | ||||
| Torymidae | Torymus geranii | ||||
| Viridiplantae | Streptophyta | Lycopodiopsida | Selaginellales | Selaginellaceae | Selaginella moellendorffii |
| Species | tRNA Type | Total | Helenus | Not Helenus |
|---|---|---|---|---|
| Puccinia triticina | tRNA-Thr-AGT | 492 | 481 | 11 |
| (LTR-1_PTrit: 426) | ||||
| (Helenus-8_PTrit: 55) | ||||
| tRNA-Ser-TGA | 189 | 183 | 6 | |
| (Helenus-2_PTrit: 95) | ||||
| (Helenus-11_PTrit: 40) | ||||
| (Helenus-11B_PTrit: 33) | ||||
| (Helenus-9_PTrit: 11) | ||||
| tRNA-Ala-TGC | 137 | 133 | 4 | |
| (Helenus-4_PTrit: 133) | ||||
| tRNA-Pro-TGG | 128 | 123 | 5 | |
| (Helenus-5_PTrit: 123) | ||||
| Puccinia striiformis | tRNA-Ala-AGC | 163 | 147 | 16 |
| (LTR-2_PSt: 145) | ||||
| tRNA-Thr-AGT | 49 | 32 | 17 | |
| (Helenus-4_PSt: 21) | ||||
| tRNA-Ser-AGA | 44 | 28 | 16 | |
| (Helenus-9_PSt: 28) | ||||
| Puccinia graminis | tRNA-Thr-AGT | 100 | 89 | 11 |
| (Helenus-3_PGr: 58) | ||||
| (Helenus-4_PGr: 15) | ||||
| Uromyces viciae-fabae | tRNA-Thr-AGT | 119 | 112 | 7 |
| (Helenus-1_UVF: 94) | ||||
| tRNA-Sup-CTA | 109 | 109 | 0 | |
| (Helenus-1_UVF: 105) | ||||
| tRNA-Ser-AGA | 95 | 90 | 5 | |
| (Helenus-2_UVF: 88) | ||||
| Cronartium ribicola | tRNA-Ala-AGC | 258 | 251 | 7 |
| (Helenus-4_CroRib: 249) | ||||
| tRNA-Sup-CTA | 245 | 245 | 0 | |
| (Helenus-5_CroRib: 194) | ||||
| (Helenus-3_CroRib: 51) | ||||
| tRNA-Thr-AGT | 160 | 154 | 6 | |
| (Helenus-2_CroRib: 88) | ||||
| (Helenus-4_CroRib: 29) | ||||
| (Helenus-5_CroRib: 22) | ||||
| tRNA-Ser-AGA | 160 | 154 | 6 | |
| (Helenus-6_CroRib: 118) | ||||
| (Helenus-13_CroRib: 13) | ||||
| tRNA-Val-GAC | 127 | 126 | 1 | |
| (Helenus-16_CroRib: 126) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kojima, K.K. Helenus and Ajax, Two Groups of Non-Autonomous LTR Retrotransposons, Represent a New Type of Small RNA Gene-Derived Mobile Elements. Biology 2024, 13, 119. https://doi.org/10.3390/biology13020119
Kojima KK. Helenus and Ajax, Two Groups of Non-Autonomous LTR Retrotransposons, Represent a New Type of Small RNA Gene-Derived Mobile Elements. Biology. 2024; 13(2):119. https://doi.org/10.3390/biology13020119
Chicago/Turabian StyleKojima, Kenji K. 2024. "Helenus and Ajax, Two Groups of Non-Autonomous LTR Retrotransposons, Represent a New Type of Small RNA Gene-Derived Mobile Elements" Biology 13, no. 2: 119. https://doi.org/10.3390/biology13020119
APA StyleKojima, K. K. (2024). Helenus and Ajax, Two Groups of Non-Autonomous LTR Retrotransposons, Represent a New Type of Small RNA Gene-Derived Mobile Elements. Biology, 13(2), 119. https://doi.org/10.3390/biology13020119