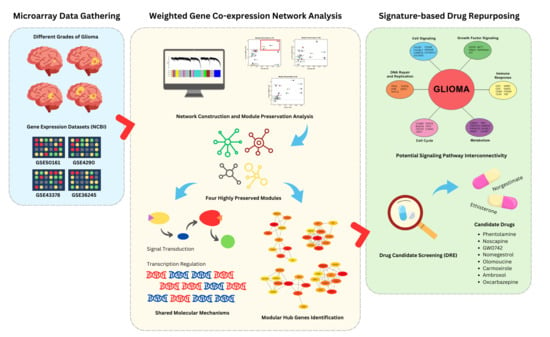
Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Acquisition and Preparation
2.2. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.2.1. Scale-Free Network Approximation
2.2.2. TOM-Based Network Construction and Module Identification
2.2.3. Module Preservation Analysis
2.3. Functional Annotation and Pathway Enrichment
2.4. Identification of Protein-Protein Interaction (PPI) and Hub Genes
2.5. Signature-Based Drug Repurposing
3. Results
3.1. Weighted Gene Co-Expression Network Analysis (WGCNA)
3.1.1. Data Pre-Processing and Approximation of Scale-Free Networks
3.1.2. TOM-Based Network Construction and Module Identification
3.2. Module Preservation Analysis
3.3. Functional Annotation and Pathway Enrichment
3.4. Identification of Protein–Protein Interaction Networks and Hub Genes
3.5. Signature-Based Drug Repurposing
4. Discussion
4.1. Gene Co-Expression Modules across the Datasets
4.2. Module Hub Genes and Their Protein Functions
4.2.1. Involvement of PI3K/Akt Pathway and Other Signaling Pathways
4.2.2. Deregulation of Cellular Processes in Glioma
4.3. Metabolic Reprogramming of Glioma Cells
4.4. Signature-Based Drug Repurposing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



| Gene | Protein | Function | Module/s |
|---|---|---|---|
| EGFR | epidermal growth factor receptor | activates MAPK and PI3K/AKT pathways; promotes cell proliferation and survival | Green, brown, and grey |
| CD8A | CD8 subunit alpha | binds to MHC class I; facilitates T cell-mediated cytotoxicity against infected or transformed cells | Green, brown, and grey |
| CALML3 | calmodulin like 3 | regulates calmodulin-dependent signaling; potential roles in intracellular calcium homeostasis | Green and brown |
| PRKACA | protein kinase cAMP-activated catalytic subunit alpha | phosphorylates target proteins; involved in cAMP signaling; regulates various cellular processes | Green and grey |
| CACNA1C | calcium voltage-gated channel subunit alpha1 C | mediates calcium ion influx; regulates neuronal excitability and cardiac function | Brown |
| CAMK2A | calcium/calmodulin dependent protein kinase II alpha | regulates synaptic plasticity and memory formation | Brown |
| PRKACB | protein kinase cAMP-activated catalytic subunit beta | involved in cAMP signaling; regulates various cellular processes | Brown |
| CACNG2 | calcium voltage-gated channel auxiliary subunit gamma 2 | modulates channel function; regulates calcium ion influx and neuronal excitability | Brown |
| KRAS | KRAS proto-oncogene, GTPase | activates MAPK pathway; regulates cell proliferation, survival, and differentiation | Brown |
| TNF | tumor necrosis factor | regulates inflammation, apoptosis, and immune cell functions | Brown |
| GRIN2B | glutamate ionotropic receptor NMDA type subunit 2B | mediates synaptic transmission; regulates synaptic plasticity, learning, and memory | Brown |
| ITGAM | integrin subunit alpha M | regulates immune cell adhesion, migration, and phagocytosis | Green |
| HSP90AA1 | heat shock protein 90 alpha family class A member 1 | assists in protein folding and stabilization | Green |
| GRB2 | growth factor receptor bound protein 2 | links receptor tyrosine kinases to Ras/MAPK pathway activation | Green |
| TLR4 | Toll-like receptor 4 | detects lipopolysaccharides; activates innate immune responses | Green |
| CD4 | CD4 molecule | binds to MHC class II; facilitates antigen recognition and T cell activation | Green |
| CD2 | CD2 molecule | regulates immune cell interactions; facilitates T cell adhesion and co-stimulation | Green |
| CALM1 | calmodulin 1 | modulates calcium-dependent signaling pathways; regulates cellular responses to calcium ions | Grey |
| EXO1 | exonuclease 1 | involved in DNA mismatch repair and recombination; maintains genomic stability | Grey |
| KIT | KIT proto-oncogene, receptor tyrosine kinase | activates PI3K/AKT and MAPK pathways; regulates cell proliferation, survival, and differentiation | Grey |
| CREB1 | cAMP responsive element binding protein 1 | binds to cAMP response elements; regulates gene expression in response to cAMP signaling | Grey |
| GNAS | G-protein alpha subunit | activates adenylyl cyclase; regulates cellular signaling pathways via GPCRs | Grey |
| ATM | ATM serine/threonine kinase | regulates DNA damage response; activates cell cycle checkpoints and DNA repair mechanisms | Grey |
| H3C12 | H3 clustered histone 12 | packaging and organizing of DNA into nucleosome | Grey |
| FN1 | fibronectin 1 | mediates cell adhesion and migration; regulates tissue remodeling and repair | Yellow |
| AKT1 | AKT serine/threonine kinase 1 | activates mTOR and other downstream pathways; regulates cell survival, growth, and metabolism | Yellow |
| BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | regulates chromosome alignment and segregation; ensures genomic stability during cell division | Yellow |
| CCNA2 | cyclin A2 | forms complexes with CDKs; regulates G1/S and G2/M transitions of the cell cycle | Yellow |
| CTNNB1 | catenin beta 1 | component of adherens junctions; regulates cell-cell adhesion and Wnt signaling pathway | Yellow |
| CDC20 | cell division cycle 20 | facilitates ubiquitination and degradation of cell cycle regulators; controls mitotic progression | Yellow |
| TP53 | tumor protein p53 | activates DNA repair or apoptosis in response to DNA damage; regulates cell cycle checkpoints | Yellow |
| CCNB1 | cyclin B1 | forms complexes with CDK1; regulates the G2/M transition of the cell cycle | Yellow |
| TOP2A | DNA topoisomerase II alpha | regulates DNA topology during replication and transcription; targeted in cancer therapy | Yellow |
| CDK1 | cyclin dependent kinase 1 | forms complexes with cyclins; regulates cell cycle transitions and mitotic entry | Yellow |
| Module | Term | Count | Adj. p-Value |
|---|---|---|---|
| Green | hsa01100 metabolic pathways | 194 | 1.4 × 10−3 |
| Brown | hsa01100 metabolic pathways | 172 | 1.3 × 10−2 |
| Yellow | hsa04151 PI3K/Akt signaling pathway | 59 | 6.3 × 10−4 |
| Gray | hsa04151 PI3K/Akt signaling pathway | 56 | 2.9 × 10−3 |
| Purple | hsa04020 calcium signaling pathway | 65 | 2.8 × 10−13 |
| Tan | hsa04015 Rap1 signaling pathway | 29 | 2.0 × 10−2 |
| Cyan | hsa04020 calcium signaling pathway | 51 | 3.6 × 10−7 |
| Black | hsa04020 calcium signaling pathway | 55 | 3.9 × 10−9 |
| Magenta | hsa04014 Ras signaling pathway | 34 | 9.2 × 10−4 |
| Pink | hsa04024 cAMP signaling pathway | 45 | 8.1 × 10−6 |
| Salmon | hsa04010 MAPK signaling pathway | 52 | 6.1 × 10−6 |
| Turquoise | hsa04010 MAPK signaling pathway | 38 | 1.3 × 10−2 |
| Blue | hsa04020 calcium signaling pathway | 68 | 1.9 × 10−10 |
| Gray60 | hsa04010 MAPK signaling pathway | 53 | 1.2 × 10−5 |
| Midnight Blue | hsa04151 PI3K/Akt signaling pathway | 51 | 4.2 × 10−3 |
| Red | hsa04115 p53 signaling pathway | 21 | 1.4 × 10−5 |
| Light Cyan | hsa05022 pathways of neurodegeneration-multiple diseases | 59 | 2.3 × 10−2 |
| Green-yellow | hsa03015 mRNA surveillance pathway | 31 | 8.8 × 10−9 |
| Light Yellow | hsa04310 Wnt signaling pathway | 25 | 9.8 × 10−3 |
| Light Green | hsa05200 pathways in cancer | 75 | 1.2 × 10−3 |
| Gold | hsa04072 phospholipase D signaling pathway | 26 | 5.1 × 10−3 |
| Category | Term | Count | Adj. p-Value |
|---|---|---|---|
| BP | GO:0007165 signal transduction | 150 | 7.8 × 10−5 |
| GO:0035556 intracellular signal transduction | 69 | 4.4 × 10−6 | |
| GO:0006468 protein phosphorylation | 64 | 3.1 × 10−5 | |
| GO:0006508 proteolysis | 62 | 2.5 × 10−4 | |
| GO:0007268 chemical synaptic transmission | 56 | 9.4 × 10−11 | |
| CC | GO:0005886 plasma membrane | 667 | 3.8 × 10−29 |
| GO:0016020 integral component of membrane | 588 | 4.4 × 10−12 | |
| GO:0005737 cytoplasm | 559 | 9.7 × 10−5 | |
| GO:0005829 cytosol | 543 | 2.0 × 10−4 | |
| GO:0016020 membrane | 424 | 1.9 × 10−12 | |
| MF | GO:0005515 protein binding | 1192 | 5.4 × 10−4 |
| GO:0005524 ATP binding | 180 | 1.1 × 10−4 | |
| GO:0042802 identical protein binding | 174 | 6.4 × 10−2 | |
| GO:0005509 calcium ion binding | 105 | 5.3 × 10−6 | |
| GO:0004712 protein serine/threonine/tyrosine kinase activity | 68 | 1.4 × 10−5 | |
| KEGG | hsa01100 metabolic pathways | 172 | 1.3 × 10−2 |
| hsa05200 pathways in cancer | 68 | 9.3 × 10−3 | |
| hsa05022 pathways of neurodegeneration-multiple diseases | 60 | 2.0 × 10−2 | |
| hsa04020 calcium signaling pathway | 49 | 2.1 × 10−6 | |
| hsa04010 MAPK signaling pathway | 48 | 4.3 × 10−4 |
| Category | Term | Count | Adj. p-Value |
|---|---|---|---|
| BP | GO:0007165 signal transduction | 181 | 4.7 × 10−10 |
| GO:0030154 cell differentiation | 89 | 1.1 × 10−3 | |
| GO:0007155 cell adhesion | 87 | 3.6 × 10−7 | |
| GO:0045087 innate immune response | 83 | 2.6 × 10−4 | |
| GO:0006915 apoptotic process | 75 | 4.2 × 10−3 | |
| CC | GO:0005886 plasma membrane | 719 | 1.0 × 10−37 |
| GO:0005737 cytoplasm | 630 | 4.9 × 10−11 | |
| GO:0016020 integral component of membrane | 595 | 3.3 × 10−10 | |
| GO:0005829 cytosol | 569 | 4.2 × 10−5 | |
| GO:0016020 membrane | 431 | 2.6 × 10−11 | |
| MF | GO:0005515 protein binding | 1256 | 3.3 × 10−7 |
| GO:0042802 identical protein binding | 214 | 4.2 × 10−6 | |
| GO:0005524 ATP binding | 172 | 4.2 × 10−3 | |
| GO:0005509 calcium ion binding | 118 | 1.0 × 10−8 | |
| GO:0042803 protein homodimerization activity | 93 | 2.3 × 10−3 | |
| KEGG | hsa01100 metabolic pathways | 194 | 1.4 × 10−3 |
| hsa05200 pathways in cancer | 82 | 1.6 × 10−4 | |
| hsa04010 MAPK signaling | 56 | 1.5 × 10−5 | |
| hsa04020 calcium signaling | 53 | 7.3 × 10−7 | |
| hsa04151 PI3K/Akt signaling pathway | 51 | 1.6 × 10−2 |
| Category | Term | Count | Adj. p-Value |
|---|---|---|---|
| BP | GO:0045944 positive regulation of transcription by RNA polymerase II | 160 | 3.9 × 10−6 |
| GO:0007165 signal transduction | 158 | 2.0 × 10−4 | |
| GO:0000122 negative regulation of transcription by RNA polymerase II | 116 | 9.2 × 10−3 | |
| GO:0045893 positive regulation of DNA-templated transcription | 97 | 1.6 × 10−4 | |
| GO:0007155 cell adhesion | 85 | 5.9 × 10−6 | |
| CC | GO:0005737 cytoplasm | 628 | 7.0 × 10−9 |
| GO:0005886 plasma membrane | 619 | 5.8 × 10−12 | |
| GO:0005829 cytosol | 596 | 1.3 × 10−6 | |
| GO:0005634 nucleus | 588 | 2.7 × 10−2 | |
| GO:0016020 integral component of membrane | 521 | 6.2 × 10−2 | |
| MF | GO:0005515 protein binding | 1307 | 8.6 × 10−9 |
| GO:0046872 metal ion binding | 288 | 2.3 × 10−2 | |
| GO:0005524 ATP binding | 217 | 1.1 × 10−9 | |
| GO:0005509 calcium ion binding | 110 | 5.5 × 10−6 | |
| GO:0004712 protein serine/threonine/tyrosine kinase activity | 78 | 1.3 × 10−7 | |
| KEGG | hsa04151 PI3K/Akt signaling pathway | 56 | 2.9 × 10−3 |
| hsa04010 MAPK signaling pathway | 49 | 7.3 × 10−4 | |
| hsa04020 calcium signaling pathway | 48 | 2.0 × 10−5 | |
| hsa04015 Rap1 signaling pathway | 38 | 4.4 × 10−4 | |
| hsa04024 cAMP signaling pathway | 36 | 5.5 × 10−3 |
| Category | Term | Count | Adj. p-Value |
|---|---|---|---|
| BP | GO:0045944 positive regulation of transcription by RNA polymerase II | 155 | 2.3 × 10−6 |
| GO:0000122 negative regulation of transcription by RNA polymerase II | 120 | 4.8 × 10−4 | |
| GO:0045893 positive regulation of DNA-templated transcription | 103 | 9.5 × 10−7 | |
| GO:0006355 regulation of DNA-templated transcription | 103 | 6.9 × 10−2 | |
| GO:0006915 apoptotic process | 99 | 4.0 × 10−9 | |
| CC | GO:0005829 cytosol | 681 | 3.1 × 10−29 |
| GO:0005634 nucleus | 679 | 7.3 × 10−19 | |
| GO:0005737 cytoplasm | 623 | 1.5 × 10−13 | |
| GO:0005654 nucleoplasm | 554 | 8.4 × 10−36 | |
| GO:0016020 membrane | 475 | 1.1 × 10−24 | |
| MF | GO:0005515 protein binding | 1396 | 3.1 × 10−46 |
| GO:0046872 metal ion binding | 268 | 7.0 × 10−2 | |
| GO:0005524 ATP binding | 232 | 1.3 × 10−15 | |
| GO:0042802 identical protein binding | 227 | 4.2 × 10−9 | |
| GO:0003723 RNA binding | 223 | 5.1 × 10−15 | |
| KEGG | hsa04151 PI3K/Akt signaling pathway | 59 | 6.3 × 10−4 |
| hsa05200 pathways in cancer | 53 | 2.7 × 10−3 | |
| hsa04141 protein processing in endoplasmic reticulum | 48 | 3.1 × 10−10 | |
| hsa05205 proteoglycans in cancer | 44 | 7.7 × 10−6 | |
| hsa04010 MAPK signaling pathway | 44 | 2.5 × 10−2 |
References
- de Almeida Sassi, F.; Lunardi Brunetto, A.; Schwartsmann, G.; Roesler, R.; Abujamra, A.L. Glioma Revisited: From Neurogenesis and Cancer Stem Cells to the Epigenetic Regulation of the Niche. J. Oncol. 2012, 2012, 537861. [Google Scholar] [CrossRef]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System Tumours: A Practical Update on What Neurosurgeons Need to Know—A Minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- van den Bent, M.J.; Brandes, A.A.; Taphoorn, M.J.B.; Kros, J.M.; Kouwenhoven, M.C.M.; Delattre, J.-Y.; Bernsen, H.J.J.A.; Frenay, M.; Tijssen, C.C.; Grisold, W.; et al. Adjuvant Procarbazine, Lomustine, and Vincristine Chemotherapy in Newly Diagnosed Anaplastic Oligodendroglioma: Long-Term Follow-Up of EORTC Brain Tumor Group Study 26951. J. Clin. Oncol. 2012, 31, 344–350. [Google Scholar] [CrossRef]
- Johnson, D.R.; Diehn, F.E.; Giannini, C.; Jenkins, R.B.; Jenkins, S.M.; Parney, I.F.; Kaufmann, T.J. Genetically Defined Oligodendroglioma Is Characterized by Indistinct Tumor Borders at MRI. Am. J. Neuroradiol. 2017, 38, 678. [Google Scholar] [CrossRef]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 Mutations in Gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 345. [Google Scholar] [CrossRef]
- Carter, T.C.; Medina-Flores, R.; Lawler, B.E. Glioblastoma Treatment with Temozolomide and Bevacizumab and Overall Survival in a Rural Tertiary Healthcare Practice. BioMed Res. Int. 2018, 2018, 6204676. [Google Scholar] [CrossRef]
- Pan, P.C.; Magge, R.S. Mechanisms of EGFR Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 8471. [Google Scholar] [CrossRef]
- Liu, X.; Chen, X.; Shi, L.; Shan, Q.; Cao, Q.; Yue, C.; Li, H.; Li, S.; Wang, J.; Gao, S.; et al. The Third-Generation EGFR Inhibitor AZD9291 Overcomes Primary Resistance by Continuously Blocking ERK Signaling in Glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 219. [Google Scholar] [CrossRef]
- Nasir, S.; Nazir, S.; Hanif, R.; Javed, A. Glioblastoma Multiforme: Probing Solutions to Systemic Toxicity towards High-Dose Chemotherapy and Inflammatory Influence in Resistance against Temozolomide. Pharmaceutics 2023, 15, 687. [Google Scholar] [CrossRef]
- Xie, X.; Bao, S.; Zhao, H.; Li, L.; Fu, X. Efficacy and Safety of Bevacizumab for Treating Glioblastoma: A Systematic Review and Meta-Analysis of Phase II and III Randomized Controlled Trials. Cancer Investig. 2023, 41, 305–317. [Google Scholar] [CrossRef]
- Reardon, D.A.; Nabors, L.B.; Mason, W.P.; Perry, J.R.; Shapiro, W.; Kavan, P.; Mathieu, D.; Phuphanich, S.; Cseh, A.; Fu, Y.; et al. Phase I/Randomized Phase II Study of Afatinib, an Irreversible ErbB Family Blocker, with or without Protracted Temozolomide in Adults with Recurrent Glioblastoma. Neuro Oncol. 2015, 17, 430–439. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Low, J.T.; Ostrom, Q.T.; Cioffi, G.; Neff, C.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. Primary Brain and Other Central Nervous System Tumors in the United States (2014–2018): A Summary of the CBTRUS Statistical Report for Clinicians. Neuro-Oncol. Pract. 2022, 9, 165–182. [Google Scholar] [CrossRef]
- Liu, H.; Bebu, I.; Li, X. Microarray Probes and Probe Sets. Front. Biosci. (Elite Ed.) 2010, 2, 325–338. [Google Scholar] [CrossRef]
- Junet, V.; Farrés, J.; Mas, J.M.; Daura, X. CuBlock: A Cross-Platform Normalization Method for gene-Expression Microarrays. Bioinformatics 2021, 37, 2365–2373. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, N.; Mai, Y.; Ren, L.; Chen, Q.; Cao, Z.; Chen, Q.; Liu, Y.; Hou, W.; Yang, J.; et al. Correcting Batch Effects in Large-Scale Multiomics Studies Using a Reference-Material-Based Ratio Method. Genome Biol. 2023, 24, 201. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, X.; Zu, Y. Integrated Analysis of WGCNA and Machine Learning Identified Biomarkers in Dilated Cardiomyopathy with Heart. Front. Cell. Dev. Biol. 2022, 10, 1089915. [Google Scholar] [CrossRef]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut Package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. CytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Li, X.; Pan, L.; Sanchez-Burgos, L.; Hühn, D.; Fernandez-Capetillo, O. The Drug Repurposing Encyclopedia (DRE): A Web Server for Systematic Drug Repurposing across 20 Organisms. bioRxiv 2023. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Horvath, S.; Geschwind, D.H. Divergence of Human and Mouse Brain Transcriptome Highlights Alzheimer Disease Pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 12698–12703. [Google Scholar] [CrossRef] [PubMed]
- Hutóczki, G.; Virga, J.; Birkó, Z.; Klekner, A. Novel Concepts of Glioblastoma Therapy Concerning Its Heterogeneity. Int. J. Mol. Sci. 2021, 22, 10005. [Google Scholar] [CrossRef] [PubMed]
- Mailem, R.C.; Tayo, L.L. Identification of Hub Genes and Key Pathways in TNF-α and IFN-γ Induced Cytokine Storms via Bioinformatics. In Proceedings of the 2022 10th International Conference on Bioinformatics and Computational Biology, ICBCB 2022, Hangzhou, China, 13–15 May 2022; pp. 6–12. [Google Scholar]
- Mailem, R.C.; Tayo, L.L. Drug Repurposing Using Gene Co-Expression and Module Preservation Analysis in Acute Respiratory Distress Syndrome (ARDS), Systemic Inflammatory Response Syndrome (SIRS), Sepsis, and COVID-19. Biology 2022, 11, 1827. [Google Scholar] [CrossRef] [PubMed]
- Manuel, M.T.A.; Tayo, L.L. Navigating the Gene Co-Expression Network and Drug Repurposing Opportunities for Brain Disorders Associated with Neurocognitive Impairment. Brain Sci. 2023, 13, 1564. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Jang, W.-Y.; Jung, T.-Y.; Jung, S.; Kim, K.-K.; Kim, H.-S.; Kim, E.-H.; Lee, M.-C.; Moon, K.-S.; Lee, K.-H. Recurrent Glioma with Lineage Conversion from oligodendroglioma to Astrocytoma in Two Cases. Front. Oncol. 2019, 9, 828. [Google Scholar] [CrossRef]
- Vitucci, M.; Karpinich, N.O.; Bash, R.E.; Werneke, A.M.; Schmid, R.S.; White, K.K.; McNeill, R.S.; Huff, B.; Wang, S.; Van Dyke, T.; et al. Cooperativity between MAPK and PI3K Signaling Activation Is Required for Glioblastoma Pathogenesis. Neuro Oncol. 2013, 15, 1317–1329. [Google Scholar] [CrossRef]
- Simms, B.A.; Zamponi, G.W. Neuronal Voltage-Gated Calcium Channels: Structure, Function, and Dysfunction. Neuron 2014, 82, 24–45. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.; Shao, W.; Wu, C.; Chen, Z.; To, S.T.; Li, W. Recent Advances in the Use of PI3K Inhibitors for Glioblastoma Multiforme: Current Preclinical and Clinical Development. Mol. Cancer 2017, 16, 100. [Google Scholar] [CrossRef]
- Gündüz, D.; Troidl, C.; Tanislav, C.; Rohrbach, S.; Hamm, C.; Aslam, M. Role of PI3K/Akt and MEK/ERK Signalling in cAMP/Epac-Mediated Endothelial Barrier Stabilisation. Front. Physiol. 2019, 10, 1387. [Google Scholar] [CrossRef]
- York, R.D.; Yao, H.; Dillon, T.; Ellig, C.L.; Eckert, S.P.; McCleskey, E.W.; Stork, P.J. Rap1 Mediates Sustained MAP Kinase Activation Induced by Nerve Factor. Nature 1998, 392, 622–626. [Google Scholar] [CrossRef]
- Kori, M.; Aydn, B.; Unal, S.; Arga, K.Y.; Kazan, D. Metabolic Biomarkers and Neurodegeneration: A Pathway Enrichment of Alzheimer’s Disease, Parkinson’s Disease, and amyotrophic Lateral Sclerosis. OMICS 2016, 20, 645–661. [Google Scholar] [CrossRef]
- Sahebjam, S.; McNamara, M.G.; Mason, W.P. Emerging Biomarkers in Anaplastic Oligodendroglioma: Implications for Clinical Investigation and Patient Management. CNS Oncol. 2013, 2, 351–358. [Google Scholar] [CrossRef]
- Barzegar Behrooz, A.; Talaie, Z.; Jusheghani, F.; Łos, M.J.; Klonisch, T.; Ghavami, S. Wnt and PI3K/Akt/MTOR Survival Pathways as Therapeutic Targets in Glioblastoma. Int. J. Mol. Sci. 2022, 23, 1353. [Google Scholar] [CrossRef]
- Weskamp, K.; Barmada, S.J. RNA Degradation in Neurodegenerative Disease. Adv. Neurobiol. 2018, 20, 103–142. [Google Scholar]
- Bruntz, R.C.; Lindsley, C.W.; Brown, H.A. Phospholipase D Signaling Pathways and Phosphatidic Acid as therapeutic Targets in Cancer. Pharmacol. Rev. 2014, 66, 1033–1079. [Google Scholar] [CrossRef]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel Mutations Target Distinct Subgroups of Medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef]
- Westphal, M.; Maire, C.L.; Lamszus, K. EGFR as a Target for Glioblastoma Treatment: An Unfulfilled Promise. CNS Drugs 2017, 31, 723–735. [Google Scholar] [CrossRef]
- Kizilbash, S.H. Why Has Targeting EGFR Aberrations in Glioblastoma Therapy Had Limited Success? Expert. Rev. Anticancer. Ther. 2022, 22, 1261–1263. [Google Scholar] [CrossRef]
- Schlegel, J.; Piontek, G.; Budde, B.; Neff, F.; Kraus, A. The Akt/Protein Kinase B-Dependent Anti-Apoptotic Pathway and the Mitogen-Activated Protein Kinase Cascade Are Alternatively Activated in Human Glioblastoma Multiforme. Cancer Lett. 2000, 158, 103–108. [Google Scholar] [CrossRef]
- So, J.-S.; Kim, H.; Han, K.-S. Mechanisms of Invasion in Glioblastoma: Extracellular Matrix, Ca2+ Signaling, and Glutamate. Front. Cell. Neurosci. 2021, 15, 663092. [Google Scholar] [CrossRef]
- Li, L.; Guan, Y.; Chen, X.; Yang, J.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Li, B.; Wu, Z.; Wu, Y.; Wang, Y.; Jin, F.; Li, D.; Ma, H.; Wang, D. Silencing of Ataxia-Telangiectasia Mutated by SiRNA Enhances the in Vitro and in Vivo Radiosensitivity of Glioma. Oncol. Rep. 2016, 35, 3303–3312. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of P53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Garcia, J.H.; Jain, S.; Aghi, M.K. Metabolic Drivers of Invasion in Glioblastoma. Front. Cell. Dev. Biol. 2021, 9, 683276. [Google Scholar] [CrossRef]
- Agnihotri, S.; Zadeh, G. Metabolic Reprogramming in Glioblastoma: The Influence of Cancer Metabolism on Epigenetics and Unanswered Questions. Neuro Oncol. 2016, 18, 160–172. [Google Scholar] [CrossRef]
- Strickland, M.; Stoll, E.A. Metabolic Reprogramming in Glioma. Front. Cell. Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef]
- Lin, H.; Patel, S.; Affleck, V.S.; Wilson, I.; Turnbull, D.M.; Joshi, A.R.; Maxwell, R.; Stoll, E.A. Fatty Acid Oxidation Is Required for the Respiration and Proliferation of Malignant Glioma Cells. Neuro Oncol. 2017, 19, 43–54. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Carrera, A.R.M.; Eleazar, E.G.; Caparanga, A.R.; Tayo, L.L. Theoretical Studies on the Quantitative Structure–Toxicity Relationship of Polychlorinated Biphenyl Congeners Reveal High Affinity Binding to Multiple Human Nuclear Receptors. Toxics 2024, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Hahn, D.W.; Allen, G.O.; McGuire, J.L. The Pharmacological Profile of Norgestimate, a New Orally Active Progestin. Contraception 1977, 16, 541–553. [Google Scholar] [CrossRef]
- McRobb, L.; Handelsman, D.J.; Kazlauskas, R.; Wilkinson, S.; McLeod, M.D.; Heather, A.K. Structure–Activity Relationships of Synthetic Progestins in a Yeast-Based in Vitro Androgen Bioassay. J. Steroid Biochem. Mol. Biol. 2008, 110, 39–47. [Google Scholar] [CrossRef]
- Huang, K.; Whelan, E.A.; Ruder, A.M.; Ward, E.M.; Deddens, J.A.; Davis-King, K.E.; Carreón, T.; Waters, M.A.; Butler, M.A.; Calvert, G.M.; et al. Reproductive Factors and Risk of Glioma in Women. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1583–1588. [Google Scholar] [CrossRef]
- McKinley, B.P.; Michalek, A.M.; Fenstermaker, R.A.; Plunkett, R.J. The Impact of Age and Gender on the Incidence of Glial Tumors in New York State from 1976–1995. J. Neurosurg. 2000, 93, 932–939. [Google Scholar] [CrossRef]
- Le Rhun, E.; Weller, M. Sex-Specific Aspects of Epidemiology, Molecular Genetics and outcome: Primary Brain Tumours. ESMO Open 2020, 5, e001034. [Google Scholar] [CrossRef] [PubMed]
- Skinner, D.C.; Evans, N.P.; Delaleu, B.; Goodman, R.L.; Bouchard, P.; Caraty, A. The Negative Feedback Actions of Progesterone On-Releasing Hormone Secretion Are Transduced by the classical Progesterone Receptor. Proc. Natl. Acad. Sci. USA 1998, 95, 10978–10983. [Google Scholar] [CrossRef] [PubMed]
- Altinoz, M.A.; Ozpinar, A.; Elmaci, I. Reproductive Epidemiology of Glial Tumors May Reveal Novel: High-Dose Progestins or Progesterone Antagonists as endocrino-Immune Modifiers against Glioma. Neurosurg. Rev. 2019, 42, 351–369. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yeung, W.L.; Zhang, P.D.; Li, N.; Kiang, M.Y.; Leung, G.K.K. Progesterone Is More Effective Than Dexamethasone in Prolonging Overall Survival and Preserving Neurologic Function in Experimental Animals with Orthotopic Glioblastoma Allografts. World Neurosurg. 2019, 125, e497–e507. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Ucal, Y.; Yilmaz, M.C.; Kiris, İ.; Ozisik, O.; Sezerman, U.; Ozpinar, A.; Elmaci, İ. Progesterone at High Doses Reduces the Growth of U87 and A172 Glioblastoma Cells: Proteomic Changes Regarding and Immunity. Cancer Med. 2020, 9, 5767–5780. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Yilmaz, M.; Ucal, Y.; Ozpinar, A.; Baskan, O.; Elmaci, I. P04.54 High Dose Progesterone Induces Growth Inhibition in Human U87 and A172 Glioblastoma Cell with Concomitant Changes in Mitochondrial and Cytoskeleton Proteins. Neuro Oncol. 2018, 20, iii291–iii292. [Google Scholar] [CrossRef]
- Germán-Castelán, L.; Manjarrez-Marmolejo, J.; González-Arenas, A.; González-Morán, M.G.; Camacho-Arroyo, I. Progesterone Induces the Growth and Infiltration of Human Astrocytoma Cells Implanted in the Cerebral Cortex of the Rat. BioMed Res. Int. 2014, 2014, 393174. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Muñoz, E.; Hernández-Hernández, O.T.; Camacho-Arroyo, I. Role of Progesterone in Human Astrocytomas Growth. Curr. Top. Med. Chem. 2011, 11, 1663–1667. [Google Scholar] [PubMed]
- Hernández-Hernández, O.T.; Camacho-Arroyo, I. Regulation of Gene Expression by Progesterone in Cancer Cells: Effects on Cyclin D1, EGFR and VEGF. Mini Rev. Med. Chem. 2013, 13, 635–642. [Google Scholar] [CrossRef]
- Atif, F.; Yousuf, S.; Stein, D.G. Anti-Tumor Effects of Progesterone in Human Glioblastoma Multiforme: Role of PI3K/Akt/MTOR Signaling. J. Steroid Biochem. Mol. Biol. 2015, 146, 62–73. [Google Scholar] [CrossRef]
- Atif, F.; Yousuf, S.; Espinosa-Garcia, C.; Sergeeva, E.; Stein, D.G. Progesterone Treatment Attenuates Glycolytic Metabolism and Induces Senescence in Glioblastoma. Sci. Rep. 2019, 9, 988. [Google Scholar] [CrossRef]
- Migliaccio, A.; Piccolo, D.; Castoria, G.; Di Domenico, M.; Bilancio, A.; Lombardi, M.; Gong, W.; Beato, M.; Auricchio, F. Activation of the Src/P21ras/Erk Pathway by Progesterone Receptor via Cross–Talk with Estrogen Receptor. EMBO J. 1998, 17, 2008–2018. [Google Scholar] [CrossRef]
- Lu, C.-C.; Tsai, S.-C. The Cyclic AMP-Dependent Protein Kinase A Pathway Is Involved in Progesterone Effects on Calcitonin Secretion from TT Cells. Life Sci. 2007, 81, 1411–1420. [Google Scholar] [CrossRef]
- Wang, Y.; Hanifi-Moghaddam, P.; Hanekamp, E.E.; Kloosterboer, H.J.; Franken, P.; Veldscholte, J.; van Doorn, H.C.; Ewing, P.C.; Kim, J.J.; Grootegoed, J.A.; et al. Progesterone Inhibition of Wnt/β-Catenin Signaling in Normal Endometrium and Endometrial Cancer. Clin. Cancer Res. 2009, 15, 5784–5793. [Google Scholar] [CrossRef]
- Luoma, J.I.; Stern, C.M.; Mermelstein, P.G. Progesterone Inhibition of Neuronal Calcium Signaling Underlies Aspects of Progesterone-Mediated Neuroprotection. J. Steroid Biochem. Mol. Biol. 2012, 131, 30–36. [Google Scholar] [CrossRef]
- Schuller, H.M. Beta-Adrenergic Signaling, a Novel Target for Cancer Therapy? Oncotarget 2010, 1, 466–469. [Google Scholar] [CrossRef]
- Braadland, P.R.; Ramberg, H.; Grytli, H.H.; Taskén, K.A. Î2-Adrenergic Receptor Signaling in Prostate Cancer. Front. Oncol. 2014, 4, 375. [Google Scholar] [CrossRef]
- Chi, T.; Wang, M.; Wang, X.; Yang, K.; Xie, F.; Liao, Z.; Wei, P. PPAR-γ Modulators as Current and Potential Cancer. Front. Oncol. 2021, 11, 737776. [Google Scholar] [CrossRef]
- Melbourne, J.K.; Thompson, K.R.; Peng, H.; Nixon, K. Chapter Eight—Its Complicated: The Relationship between Alcohol and Microglia in the Search for Novel Pharmacotherapeutic Targets for Alcohol Use Disorders. In Molecular Basis of Neuropsychiatric Disorders: From Bench to Bedside; Rahman, S., Ed.; Progress in Molecular Biology and Translational Science; Academic Press: Cambridge, MA, USA, 2019; Volume 167, pp. 179–221. [Google Scholar]
- Pucci, B.; Kasten, M.; Giordano, A. Cell Cycle and Apoptosis. Neoplasia 2000, 2, 291–299. [Google Scholar] [CrossRef]
- Griffin, M.; Khan, R.; Basu, S.; Smith, S. Ion Channels as Therapeutic Targets in High Grade Gliomas. Cancers 2020, 12, 3068. [Google Scholar] [CrossRef]
- Seifert, S.; Sontheimer, H. Bradykinin Enhances Invasion of Malignant Glioma into the Brain Parenchyma by Inducing Cells to Undergo Amoeboid Migration. J. Physiol. 2014, 592, 5109–5127. [Google Scholar] [CrossRef]
- Yang, K.; Wei, M.; Yang, Z.; Fu, Z.; Xu, R.; Cheng, C.; Chen, X.; Chen, S.; Dammer, E.; Le, W. Activation of Dopamine Receptor D1 Inhibits Glioblastoma Tumorigenicity by Regulating Autophagic Activity. Cell. Oncol. 2020, 43, 1175–1190. [Google Scholar] [CrossRef]
- Jeon, H.-M.; Oh, Y.T.; Shin, Y.J.; Chang, N.; Kim, D.; Woo, D.; Yeup, Y.; Joo, K.M.; Jo, H.; Yang, H.; et al. Dopamine Receptor D2 Regulates Glioblastoma Survival and death through MET and Death Receptor 4/5. Neoplasia 2023, 39, 100894. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Harris, M.A.; Huntley, R.; Van Auken, K.; Cherry, J.M. A Guide to Best Practices for Gene Ontology (GO) Manual Annotation. Database 2013, 2013, bat054. [Google Scholar] [CrossRef] [PubMed]








| Accession No. | GSE50161 | GSE4290 | GSE43378 | GSE36245 |
|---|---|---|---|---|
| Condition | Pilocytic Astrocytoma | Oligodendroglioma | Anaplastic Astrocytoma | Glioblastoma Multiforme |
| Type | Expression Profiling by Array | |||
| Platform | GPL570-HG-U133 Plus 2 Affymetrix Human Genome U133 Plus 2.0 Array | |||
| Source | Primary Brain Tumor Samples | |||
| No. of Samples | 14 | 36 | 19 | 44 |
| Expression | Genes | Drug | Mechanism | Tau | FDR |
|---|---|---|---|---|---|
| Upregulated | KRAS, CCNB1, BUB1B, KIT, TP53, EGFR, ATM, EXO1, GNAS, CDC20, TOP2A, HSP90AA1, FN1, H3C12, GRIN2B, GRB2, CCNA2, CDK1, CALM1, CALML3, CAMK2A, CREB1, TNF, AKT1, CTNNB1, and ITGAM | Norgestimate Phentolamine GW0742 Olomoucine Ambroxol | Progesterone receptor agonist Adrenergic receptor antagonist PPAR receptor agonist CDK inhibitor Sodium channel blocker | −99.8 −99.7 −99.5 −99.4 −99.3 | 7.75 × 10−3 6.53 × 10−3 5.05 × 10−4 7.01 × 10−3 8.35 × 10−3 |
| Downregulated | CD4, CACNA1C, CACNG2, CD2, PRKACA, PRKACB, TLR4, and CD8A | Ethisterone Noscapine Nomegestrol Carmoxirole Oxcarbazepine | Progestogen hormone Bradykinin receptor antagonist Progestogen hormone Dopamine receptor agonist Sodium channel blocker | −99.7 −99.6 −99.5 −99.5 −99.5 | 2.36 × 10−3 6.22 × 10−5 7.89 × 10−4 7.40 × 10−3 9.94 × 10−3 |
| Drug | Status | Pathway/Process | Reference |
|---|---|---|---|
| Norgestimate | Approved | PI3K/Akt Pathway | [68,69,70] |
| Phentolamine | Approved | cAMP Signaling, MAPK/ERK, and PI3K/Akt Pathway | [75,76] |
| GW0742 | Experimental | Lipid Metabolic Pathways | [77,78] |
| Olomoucine | Approved | Cell Cycle | [79] |
| Ambroxol | Approved | Calcium Signaling Pathway | [80] |
| Ethisterone | Approved | PI3K/Akt Pathway | [68,69,70] |
| Noscapine | Approved | Calcium Signaling Pathway | [81] |
| Nomegestrol | Approved | PI3K/Akt Pathway | [68,69,70] |
| Carmoxirole | Experimental | cAMP Signaling Pathway | [82,83] |
| Oxcarbazepine | Approved | Calcium Signaling Pathway | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orda, M.A.; Fowler, P.M.P.T.; Tayo, L.L. Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades. Biology 2024, 13, 206. https://doi.org/10.3390/biology13040206
Orda MA, Fowler PMPT, Tayo LL. Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades. Biology. 2024; 13(4):206. https://doi.org/10.3390/biology13040206
Chicago/Turabian StyleOrda, Marco A., Peter Matthew Paul T. Fowler, and Lemmuel L. Tayo. 2024. "Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades" Biology 13, no. 4: 206. https://doi.org/10.3390/biology13040206
APA StyleOrda, M. A., Fowler, P. M. P. T., & Tayo, L. L. (2024). Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades. Biology, 13(4), 206. https://doi.org/10.3390/biology13040206