Polymorphism Analysis of pfmdr1 and pfcrt from Plasmodium falciparum Isolates in Northwestern Nigeria Revealed the Major Markers Associated with Antimalarial Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site, Samples and Ethics
2.2. DNA Extraction and Confirmation of P. falciparum Infection Using PCR
2.3. Amplification of Pfmdr1 Fragment Encompassing the 86th Codon
2.4. Genotyping of the Pfmdr1 N86Y Mutation Using Restriction Fragment Length Polymorphism (RFLP)
2.5. Amplification, Cloning and Sequencing of Pfmdr1 Fragment Encompassing 86th and 184th Codons
2.6. Amplification, Cloning and Sequencing of Pfcrt Fragment Encompassing the 76th Codon
2.7. Data Analysis
3. Results
3.1. Cytochrome Oxidase III PCR Confirmation of Infection
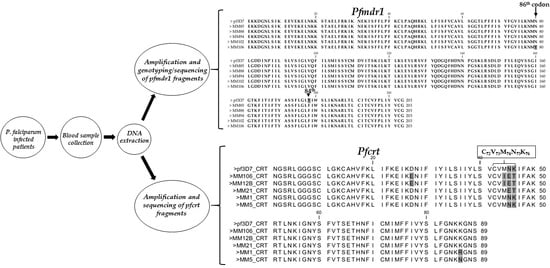
3.2. Amplification of Pfmdr1 Fragment Encompassing the 86th Codon and Genotyping
3.3. Polymorphism Analysis of Fragments of Pfmdr1 and Pfcrt
4. Discussion
4.1. Evidence of Resistance Haplotypes in pfmdr1 in P. falciparum Isolates from Kano
4.2. Evidence of Resistance Haplotypes in pfcrt in P. falciparum Isolates from Kano
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report 2019; 9241565721; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Bhatt, S.; Weiss, D.; Cameron, E.; Bisanzio, D.; Mappin, B.; Dalrymple, U.; Battle, K.; Moyes, C.; Henry, A.; Eckhoff, P. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, P.; Noor, A.M. The global fight against malaria is at crossroads. Lancet 2017, 390, 2532–2534. [Google Scholar] [CrossRef]
- Shrivastava, S.K.; Gupta, R.K.; Mahanta, J.; Dubey, M.L. Correlation of molecular markers, Pfmdr1-N86Y and Pfcrt-K76T, with in vitro chloroquine resistant Plasmodium falciparum, isolated in the malaria endemic states of Assam and Arunachal Pradesh, Northeast India. PLoS ONE 2014, 9, e103848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dondorp, A.M.; Nosten, F.; Yi, P.; Das, D.; Phyo, A.P.; Tarning, J.; Lwin, K.M.; Ariey, F.; Hanpithakpong, W.; Lee, S.J.; et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009, 361, 455–467. [Google Scholar] [CrossRef] [Green Version]
- WHO. Artemisinin and Artemisinin-based Combination Therapy Resistance: Status Report; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [Green Version]
- MalariaGEN. Genomic Epidemiology of Artemisinin Resistant Malaria; MalariaGEN: Oxford, UK, 2016. [Google Scholar]
- WWARN Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019, 17, 1. [Google Scholar] [CrossRef] [Green Version]
- Duraisingh, M.T.; Cowman, A.F. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005, 94, 181–190. [Google Scholar] [CrossRef]
- Folarin, O.A.; Bustamante, C.; Gbotosho, G.O.; Sowunmi, A.; Zalis, M.G.; Oduola, A.M.; Happi, C.T. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011, 120, 224–230. [Google Scholar] [CrossRef] [Green Version]
- Buppan, P.; Seethamchai, S.; Kuamsab, N.; Harnyuttanakorn, P.; Putaporntip, C.; Jongwutiwes, S. Multiple Novel Mutations in Plasmodium falciparum Chloroquine Resistance Transporter Gene during Implementation of Artemisinin Combination Therapy in Thailand. Am. J. Trop. Med. Hyg. 2018, 99, 987–994. [Google Scholar] [CrossRef]
- Happi, C.T.; Gbotosho, G.O.; Folarin, O.A.; Bolaji, O.M.; Sowunmi, A.; Kyle, D.E.; Milhous, W.; Wirth, D.F.; Oduola, A.M. Association between mutations in Plasmodium falciparum chloroquine resistance transporter and P. falciparum multidrug resistance 1 genes and in vivo amodiaquine resistance in P. falciparum malaria-infected children in Nigeria. Am. J. Trop. Med. Hyg. 2006, 75, 155–161. [Google Scholar] [CrossRef]
- FMoH. National Antimalarial Treatment Guidelines Policy; Federal Ministry of Health National Malaria and Vector Control Division: Abuja, Nigeria, 2005.
- Ajayi, N.A.; Ukwaja, K.N. Possible artemisinin-based combination therapy-resistant malaria in Nigeria: A report of three cases. Rev. Soc. Bras. Med. Trop. 2013, 46, 525–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wundermann, G.; Osiki, A. Currently Observed Trend in the Resistance of Malaria to Artemisinin Based Combination Therapy in Nigeria–A Report of 5 Cases. Int. J. Trop. Dis. Health 2017, 21, 1–5. [Google Scholar] [CrossRef]
- Bustamante, C.; Folarin, O.A.; Gbotosho, G.O.; Batista, C.N.; Mesquita, E.A.; Brindeiro, R.M.; Tanuri, A.; Struchiner, C.J.; Sowunmi, A.; Oduola, A.; et al. In vitro-reduced susceptibility to artemether in P. falciparum and its association with polymorphisms on transporter genes. J. Infect. Dis. 2012, 206, 324–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oboh, M.A.; Ndiaye, D.; Antony, H.A.; Badiane, A.S.; Singh, U.S.; Ali, N.A.; Bharti, P.K.; Das, A. Status of Artemisinin Resistance in Malaria Parasite Plasmodium falciparum from Molecular Analyses of the Kelch13 Gene in Southwestern Nigeria. Biomed. Res. Int. 2018, 2018, 2305062. [Google Scholar] [CrossRef] [Green Version]
- Igbasi, U.; Oyibo, W.; Omilabu, S.; Quan, H.; Chen, S.B.; Shen, H.M.; Chen, J.H.; Zhou, X.N. Kelch 13 propeller gene polymorphism among Plasmodium falciparum isolates in Lagos, Nigeria: Molecular Epidemiologic Study. Trop. Med. Int. Health. 2019, 24, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Dokunmu, T.M.; Olasehinde, G.I.; Oladejo, D.O.; Adjekukor, C.U.; Akinbohun, A.E.; Onileere, O.; Eze, C.J.; Jir, G.S. Evaluation of Plasmodium falciparum K13 gene polymorphism and susceptibility to dihydroartemisinin in an endemic area. Biomed. Res. Ther. 2018, 5, 2651–2657. [Google Scholar] [CrossRef]
- Idowu, A.O.; Oyibo, W.A.; Bhattacharyya, S.; Khubbar, M.; Mendie, U.E.; Bumah, V.V.; Black, C.; Igietseme, J.; Azenabor, A.A. Rare mutations in Pfmdr1 gene of Plasmodium falciparum detected in clinical isolates from patients treated with anti-malarial drug in Nigeria. Malar. J. 2019, 18, 319. [Google Scholar] [CrossRef]
- Abubakar, U.F.; Adam, R.; Mukhtar, M.M.; Muhammad, A.; Yahuza, A.A.; Ibrahim, S.S. Identification of Mutations in Antimalarial Resistance Gene Kelch13 from plasmodium falciparum Isolates in Kano, Nigeria. Trop. Med. Infect. Dis. 2020, 5. [Google Scholar] [CrossRef]
- Muhammad, R.H.; Nock, I.H.; Ndams, I.S.; George, J.B.; Deeni, Y. Distribution of Pfmdr1 and Pfcrt chloroquine drug resistance alleles in north-western Nigeria. MWJ 2017, 8, 15. [Google Scholar]
- Ikegbunam, M.N.; Nkonganyi, C.N.; Thomas, B.N.; Esimone, C.O.; Velavan, T.P.; Ojurongbe, O. Analysis of Plasmodium falciparum Pfcrt and Pfmdr1 genes in parasite isolates from asymptomatic individuals in Southeast Nigeria 11 years after withdrawal of chloroquine. Malar. J. 2019, 18, 343. [Google Scholar] [CrossRef] [Green Version]
- Agomo, C.O.; Oyibo, W.A.; Sutherland, C.; Hallet, R.; Oguike, M. Assessment of Markers of Antimalarial Drug Resistance in Plasmodium falciparum Isolates from Pregnant Women in Lagos, Nigeria. PLoS ONE 2016, 11, e0146908. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.B.; Saliba, K.J.; Caruana, S.R.; Kirk, K.; Cowman, A.F. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 2000, 403, 906–909. [Google Scholar] [CrossRef] [PubMed]
- Duraisingh, M.T.; Jones, P.; Sambou, I.; Von Seidlein, L.; Pinder, M.; Warhurst, D.C. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 2000, 108, 13–23. [Google Scholar] [CrossRef]
- Veiga, M.I.; Dhingra, S.K.; Henrich, P.P.; Straimer, J.; Gnadig, N.; Uhlemann, A.C.; Martin, R.E.; Lehane, A.M.; Fidock, D.A. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016, 7, 11553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menard, D.; Dondorp, A. Antimalarial Drug Resistance: A Threat to Malaria Elimination. Cold Spring Harb. Perspect Med. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Dokunmu, T.M.; Adjekukor, C.U.; Yakubu, O.F.; Bello, A.O.; Adekoya, J.O.; Akinola, O.; Amoo, E.O.; Adebayo, A.H. Asymptomatic malaria infections and Pfmdr1 mutations in an endemic area of Nigeria. Malar. J. 2019, 18, 218. [Google Scholar] [CrossRef] [Green Version]
- Fidock, D.A.; Nomura, T.; Talley, A.K.; Cooper, R.A.; Dzekunov, S.M.; Ferdig, M.T.; Ursos, L.M.; Sidhu, A.B.; Naude, B.; Deitsch, K.W.; et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 2000, 6, 861–871. [Google Scholar] [CrossRef]
- Djimde, A.; Doumbo, O.K.; Cortese, J.F.; Kayentao, K.; Doumbo, S.; Diourte, Y.; Coulibaly, D.; Dicko, A.; Su, X.Z.; Nomura, T.; et al. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001, 344, 257–263. [Google Scholar] [CrossRef]
- Awasthi, G.; Satya Prasad, G.B.; Das, A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem. Inst. Oswaldo Cruz. 2012, 107, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Ariey, F.; Fandeur, T.; Durand, R.; Randrianarivelojosia, M.; Jambou, R.; Legrand, E.; Ekala, M.T.; Bouchier, C.; Cojean, S.; Duchemin, J.B.; et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar. J. 2006, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Echeverry, D.F.; Deason, N.A.; Davidson, J.; Makuru, V.; Xiao, H.; Niedbalski, J.; Kern, M.; Russell, T.L.; Burkot, T.R.; Collins, F.H.; et al. Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene. Malar. J. 2016, 15, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basco, L.K.; Ringwald, P. Molecular epidemiology of malaria in Cameroon. X. Evaluation of PFMDR1 mutations as genetic markers for resistance to amino alcohols and artemisinin derivatives. Am. J. Trop. Med. Hyg. 2002, 66, 667–671. [Google Scholar] [CrossRef] [Green Version]
- Goldfless, S.J.; Wagner, J.C.; Niles, J.C. Versatile control of Plasmodium falciparum gene expression with an inducible protein-RNA interaction. Nat. Commun. 2014, 5, 5329. [Google Scholar] [CrossRef] [Green Version]
- Payne, D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol. Today 1987, 3, 241–246. [Google Scholar] [CrossRef]
- Brieger, W. Where is Chloroquine Now? Afr. Health 2017, 39, 13–15. [Google Scholar]
- Wurtz, N.; Fall, B.; Pascual, A.; Fall, M.; Baret, E.; Camara, C.; Nakoulima, A.; Diatta, B.; Fall, K.B.; Mbaye, P.S.; et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob. Agents Chemother. 2014, 58, 7032–7040. [Google Scholar] [CrossRef] [Green Version]
- Oladipo, O.O.; Wellington, O.A.; Sutherland, C.J. Persistence of chloroquine-resistant haplotypes of Plasmodium falciparum in children with uncomplicated Malaria in Lagos, Nigeria, four years after change of chloroquine as first-line antimalarial medicine. Diagn. Pathol. 2015, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Sisowath, C.; Ferreira, P.E.; Bustamante, L.Y.; Dahlstrom, S.; Martensson, A.; Bjorkman, A.; Krishna, S.; Gil, J.P. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop. Med. Int. Health 2007, 12, 736–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laufer, M.K.; Plowe, C.V. Withdrawing antimalarial drugs: Impact on parasite resistance and implications for malaria treatment policies. Drug Resist. Updat. 2004, 7, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Laufer, M.K.; Thesing, P.C.; Eddington, N.D.; Masonga, R.; Dzinjalamala, F.K.; Takala, S.L.; Taylor, T.E.; Plowe, C.V. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 2006, 355, 1959–1966. [Google Scholar] [CrossRef]
- Ibraheem, Z.O.; Abd Majid, R.; Noor, S.M.; Sedik, H.M.; Basir, R. Role of Different Pfcrt and Pfmdr-1 Mutations in Conferring Resistance to Antimalaria Drugs in Plasmodium falciparum. Malar. Res. Treat. 2014, 2014, 950424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Wei, Q.; Yin, K.; Sun, H.; Li, J.; Xiao, T.; Kong, X.; Wang, Y.; Zhao, G.; Zhu, S.; et al. Surveillance of Antimalarial Resistance Pfcrt, Pfmdr1, and Pfkelch13 Polymorphisms in African Plasmodium falciparum imported to Shandong Province, China. Sci. Rep. 2018, 8, 12951. [Google Scholar] [CrossRef]




| Location | N | S | H | Hd | Syn | Nonsyn | π (k) | D (Tajima) | D* (Fu and Li) |
|---|---|---|---|---|---|---|---|---|---|
| pfmdr1 | |||||||||
| Kano | 16 | 2 | 3 | 0.49 | 0 | 2 | 0.00096 (0.583) | −0.0823 ns | −0.5038 ns |
| pfcrt | |||||||||
| Kano | 18 | 7 | 6 | 0.72 | 0 | 7 | 0.00879 (2.346) | 0.518 ns | 0.0939 ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, R.; Mukhtar, M.M.; Abubakar, U.F.; Damudi, H.A.; Muhammad, A.; Ibrahim, S.S. Polymorphism Analysis of pfmdr1 and pfcrt from Plasmodium falciparum Isolates in Northwestern Nigeria Revealed the Major Markers Associated with Antimalarial Resistance. Diseases 2021, 9, 6. https://doi.org/10.3390/diseases9010006
Adam R, Mukhtar MM, Abubakar UF, Damudi HA, Muhammad A, Ibrahim SS. Polymorphism Analysis of pfmdr1 and pfcrt from Plasmodium falciparum Isolates in Northwestern Nigeria Revealed the Major Markers Associated with Antimalarial Resistance. Diseases. 2021; 9(1):6. https://doi.org/10.3390/diseases9010006
Chicago/Turabian StyleAdam, Ruqayya, Muhammad M. Mukhtar, Umar F. Abubakar, Hajara A. Damudi, Abdullahi Muhammad, and Sulaiman S. Ibrahim. 2021. "Polymorphism Analysis of pfmdr1 and pfcrt from Plasmodium falciparum Isolates in Northwestern Nigeria Revealed the Major Markers Associated with Antimalarial Resistance" Diseases 9, no. 1: 6. https://doi.org/10.3390/diseases9010006
APA StyleAdam, R., Mukhtar, M. M., Abubakar, U. F., Damudi, H. A., Muhammad, A., & Ibrahim, S. S. (2021). Polymorphism Analysis of pfmdr1 and pfcrt from Plasmodium falciparum Isolates in Northwestern Nigeria Revealed the Major Markers Associated with Antimalarial Resistance. Diseases, 9(1), 6. https://doi.org/10.3390/diseases9010006