Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach
Abstract
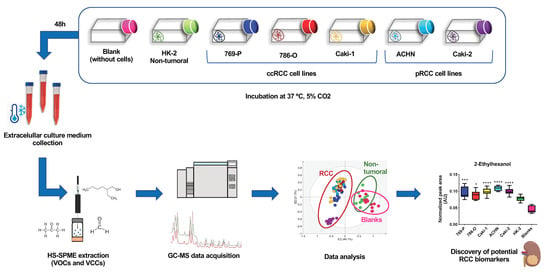
:1. Introduction
2. Results
2.1. Volatile Exometabolome Signature of RCC Cell Lines versus the Non-Tumorigenic Cell Line
2.2. Volatile Exometabolome Signature of Metastatic versus Non-Metastatic RCC Cell Lines and ccRCC versus pRCC Cell Lines
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell lines and Culture Conditions
4.3. Sample Preparation and Volatile Extraction by HS-SPME
4.4. GC–MS Analysis: Equipment and Conditions
4.5. Compound Identification and GC–MS Data Pre-Processing
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernandez-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Liu, X.; Sun, H.; Guo, Z.; Tang, X.; Wang, Z.; Li, J.; Li, H.; Sun, W.; et al. Urine Metabolomics for Renal Cell Carcinoma (RCC) Prediction: Tryptophan Metabolism as an Important Pathway in RCC. Front. Oncol. 2019, 9, 663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muglia, V.F.; Prando, A. Renal cell carcinoma: histological classification and correlation with imaging findings*. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheville, J.C.; Lohse, C.M.; Zincke, H.; Weaver, A.L.; Blute, M.L. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am. J. Surg. Pathol. 2003, 27, 612–624. [Google Scholar] [CrossRef]
- Monteiro, M.; Carvalho, M.; Bastos, M.L.; De Pinho, P.G. Biomarkers in renal cell carcinoma: A metabolomics approach. Metabolomics 2014, 10, 1210–1222. [Google Scholar] [CrossRef]
- Deng, J.; Li, L.; Xia, H.; Guo, J.; Wu, X.; Yang, X.; Hong, Y.; Chen, Q.; Hu, J. A comparison of the prognosis of papillary and clear cell renal cell carcinoma: Evidence from a meta-analysis. Medicine 2019, 98, e16309. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, J.; Zhao, L.; Chen, M.; Shi, A.; Pan, L.; Huang, Y.; Zhang, H.; Dong, B.; Gao, H. Prediction and diagnosis of renal cell carcinoma using nuclear magnetic resonance-based serum metabolomics and self-organizing maps. Oncotarget 2016, 7, 59189–59198. [Google Scholar] [CrossRef] [Green Version]
- Popławski, P.; Tohge, T.; Bogusławska, J.; Rybicka, B.; Tański, Z.; Trevino, V.; Fernie, A.R.; Piekiełko-Witkowska, A. Integrated transcriptomic and metabolomic analysis shows that disturbances in metabolism of tumor cells contribute to poor survival of RCC patients. Biochim. et Biophys. Acta (BBA) - Mol. Basis Dis. 2017, 1863, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Farber, N.J.; Kim, C.J.; Modi, P.K.; Hon, J.D.; Sadimin, E.; Singer, E.A. Renal cell carcinoma: the search for a reliable biomarker. Transl. Cancer Res. 2017, 6, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Nath, K.; Bansal, N.; Kumar, M. Role of metabolomics-derived biomarkers to identify renal cell carcinoma: a comprehensive perspective of the past ten years and advancements. Expert Rev. Mol. Diagn. 2019, 20, 5–18. [Google Scholar] [CrossRef]
- Weiss, R.; Lin, P.-Y. Kidney cancer: Identification of novel targets for therapy. Kidney Int. 2006, 69, 224–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutson, T.E. Renal Cell Carcinoma: Diagnosis and Treatment, 1994–2003. Bayl. Univ. Med Cent. Proc. 2005, 18, 337–340. [Google Scholar] [CrossRef] [Green Version]
- Armitage, E.G.; Ciborowski, M. Applications of Metabolomics in Cancer Studies. Adv. Exp. Med. Biol. 2017, 965, 209–234. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Banerjee, H.N. Metabolomic Approaches in Cancer Epidemiology. Diseases 2015, 3, 167–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wettersten, H.I.; Abu Aboud, O.; Lara, P.N.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef]
- Rodrigues, D.; Monteiro, M.; Jerónimo, C.; Henrique, R.; Belo, L.; Bastos, M.L.; De Pinho, P.G.; Carvalho, M. Renal cell carcinoma: a critical analysis of metabolomic biomarkers emerging from current model systems. Transl. Res. 2017, 180, 1–11. [Google Scholar] [CrossRef]
- Nizioł, J.; Bonifay, V.; Ossoliński, K.; Ossoliński, T.; Ossolińska, A.; Sunner, J.; Beech, I.; Arendowski, A.; Ruman, T. Metabolomic study of human tissue and urine in clear cell renal carcinoma by LC-HRMS and PLS-DA. Anal. Bioanal. Chem. 2018, 410, 3859–3869. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Dong, B.; Jia, J.; Zhu, H.; Diao, C.; Yan, Z.-H.; Huang, Y.; Li, X. Application of ex vivo 1H NMR metabonomics to the characterization and possible detection of renal cell carcinoma metastases. J. Cancer Res. Clin. Oncol. 2012, 138, 753–761. [Google Scholar] [CrossRef]
- Falegan, O.S.; Ball, M.; Shaykhutdinov, R.A.; Pieroraio, P.M.; Farshidfar, F.; Vogel, H.J.; Allaf, M.E.; Hyndman, M.E. Urine and Serum Metabolomics Analyses May Distinguish between Stages of Renal Cell Carcinoma. Metabolites 2017, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, K.; Podmore, I. Current Challenges in Volatile Organic Compounds Analysis as Potential Biomarkers of Cancer. J. Biomark. 2015, 2015, 981458. [Google Scholar] [CrossRef] [Green Version]
- Lima, A.R.C.M.; Araujo, A.M.; Pinto, J.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; Carvalho, M.; De Pinho, P.G. Discrimination between the human prostate normal and cancer cell exometabolome by GC-MS. Sci. Rep. 2018, 8, 5539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Patra, A.; Kutty, V.K.; Venkatesan, V. Critical Review of Volatile Organic Compound Analysis in Breath and In Vitro Cell Culture for Detection of Lung Cancer. Metabolites 2019, 9, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Zhang, H.; Ong, C.N.; Patra, A.; Lu, Y.; Lim, C.T.; Venkatesan, T. Detection of Lung Cancer: Concomitant Volatile Organic Compounds and Metabolomic Profiling of Six Cancer Cell Lines of Different Histological Origins. ACS Omega 2018, 3, 5131–5140. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Pinto, J.; Araujo, A.M.; Reis, S.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; De Pinho, P.G.; Carvalho, M. Volatile metabolomic signature of bladder cancer cell lines based on gas chromatography–mass spectrometry. Metabolomics 2018, 14, 62. [Google Scholar] [CrossRef]
- Silva, C.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Volatile metabolomic signature of human breast cancer cell lines. Sci. Rep. 2017, 7, 43969. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, M.; Moreira, N.; Pinto, J.; Pires-Luís, A.S.; Henrique, R.; Jerónimo, C.; Bastos, M.L.; Gil, A.M.; Carvalho, M.; De Pinho, P.G. GC-MS metabolomics-based approach for the identification of a potential VOC-biomarker panel in the urine of renal cell carcinoma patients. J. Cell. Mol. Med. 2017, 21, 2092–2105. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Pi, X.; Guo, L.; Wang, Y.; Li, M.; Feng, Y.; Lin, Z.; Hou, W.; Li, E. Urinary volatile organic compounds as potential biomarkers for renal cell carcinoma. Biomed. Rep. 2016, 5, 68–72. [Google Scholar] [CrossRef] [Green Version]
- Lima, A.R.C.M.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; De Pinho, P.G.; Carvalho, M. Identification of a biomarker panel for improvement of prostate cancer diagnosis by volatile metabolic profiling of urine. Br. J. Cancer 2019, 121, 857–868. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Boil. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef]
- Ganesamoni, R.; Bhattacharyya, S.; Kumar, S.; Chauhan, A.; Mete, U.K.; Agarwal, M.M.; Mavuduru, R.; Kaushik, G.; Mandal, A.K.; Singh, S.K. Status of Oxidative Stress in Patients With Renal Cell Carcinoma. J. Urol. 2012, 187, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Shanmugasundaram, K.; Block, K. Renal Carcinogenesis, Tumor Heterogeneity, and Reactive Oxygen Species: Tactics Evolved. Antioxid. Redox Signal. 2016, 25, 685–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šverko, A.; Sobočanec, S.; Kušić, B.; Mačak-Šafranko, Ž.; Šarić, A.; Leniček, T.; Kraus, O.; Andrisic, L.; Korolija, M.; Balog, T.; et al. Superoxide dismutase and cytochrome P450 isoenzymes might be associated with higher risk of renal cell carcinoma in male patients. Int. Immunopharmacol. 2011, 11, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Gago-Dominguez, M.; Castelao, J.E. Lipid peroxidation and renal cell carcinoma: further supportive evidence and new mechanistic insights. Free. Radic. Boil. Med. 2006, 40, 721–733. [Google Scholar] [CrossRef]
- Li, H.; Li, W.-X.; Dai, S.; Guo, Y.; Zheng, J.; Liu, J.; Wang, Q.; Chen, B.; Li, G.-H.; Huang, J. Identification of metabolism-associated genes and pathways involved in different stages of clear cell renal cell carcinoma. Oncol. Lett. 2017, 15, 2316–2322. [Google Scholar] [CrossRef]
- McFadyen, M.; Melvin, W.T.; I Murray, G. Cytochrome P450 CYP1B1 activity in renal cell carcinoma. Br. J. Cancer 2004, 91, 966–971. [Google Scholar] [CrossRef] [Green Version]
- Hanai, Y.; Shimono, K.; Matsumura, K.; Vachani, A.; Albelda, S.; Yamazaki, K.; Beauchamp, G.K.; Oka, H. Urinary Volatile Compounds as Biomarkers for Lung Cancer. Biosci. Biotechnol. Biochem. 2012, 76, 679–684. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Pacheco, A.; Salinero-Bachiller, M.; Iribar, M.C.; López-Luque, A.; Miján-Ortiz, J.L.; Peinado, J.M. Furan and p-xylene as candidate biomarkers for prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 243.e21–243.e27. [Google Scholar] [CrossRef]
- Orywal, K.; Jelski, W.; Werel, T.; Szmitkowski, M. The activity of class I, II, III and IV alcohol dehydrogenase isoenzymes and aldehyde dehydrogenase in renal cell carcinoma. Exp. Mol. Pathol. 2015, 98, 403–406. [Google Scholar] [CrossRef]
- Orywal, K.; Szmitkowski, M. Alcohol dehydrogenase and aldehyde dehydrogenase in malignant neoplasms. Clin. Exp. Med. 2016, 17, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Weiss, R.H. Metabolomics and Metabolic Reprogramming in Kidney Cancer. Semin. Nephrol. 2018, 38, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Schaeffeler, E.; Büttner, F.; Reustle, A.; Klumpp, V.; Winter, S.; Rausch, S.; Fisel, P.; Hennenlotter, J.; Kruck, S.; Stenzl, A.; et al. Metabolic and Lipidomic Reprogramming in Renal Cell Carcinoma Subtypes Reflects Regions of Tumor Origin. Eur. Urol. Focus 2019, 5, 608–618. [Google Scholar] [CrossRef] [Green Version]
- Jing, L.; Guigonis, J.-M.; Borchiellini, D.; Durand, M.; Pourcher, T.; Ambrosetti, D. LC-MS based metabolomic profiling for renal cell carcinoma histologic subtypes. Sci. Rep. 2019, 9, 15635. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, A.A.; Reznik, E.; Lee, C.-H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; A Weinberg, R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the right cell line for renal cell cancer research. Mol. Cancer 2016, 15, 83. [Google Scholar] [CrossRef] [Green Version]
- Ryan, M.J.; Johnson, G.; Kirk, J.; Fuerstenberg, S.M.; Zager, R.A.; Torok-Storb, B. HK-2: An immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994, 45, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Araujo, A.M.; Moreira, N.; Lima, A.R.; Bastos, M.L.; Carvalho, F.; Carvalho, M.; De Pinho, P.G. Analysis of extracellular metabolome by HS-SPME/GC–MS: Optimization and application in a pilot study to evaluate galactosamine-induced hepatotoxicity. Toxicol. Lett. 2018, 295, 22–31. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]




| Metabolite | 769-P (n = 13) vs. HK-2 (n = 12) | 786-O (n = 14) vs. HK-2 (n = 12) | Caki-1 (n = 11) vs. HK-2 (n = 12) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ES ± SE | p-Value a | AUC | ES ± SE | p-Value a | AUC | ES ± SE | p-Value a | AUC | |||
| Alcohols | |||||||||||
| 2-Ethylhexanol b, L2, * | ↑1.42 ± 0.86 E | 1.26 × 10−3 | 0.885 | ↑0.94 ± 0.79 E | 4.88 × 10−2 | 0.738 | ↑2.39 ± 1.05 E | 6.21 × 10−4 | 0.955 | ||
| 2,6-Dimethyl-7-octen-2-ol b, L2 | ↓−1.79 ± 0.94 C | 2.02 × 10−3 | 0.916 | ||||||||
| Cyclohexanol b, L2 | ↓−1.29 ± 0.82 C | 8.06 × 10−3 | 0.821 | ||||||||
| Levomenthol b, L2 | ↓−0.84 ± 0.79 | 4.90 × 10−2 | 0.734 | ↓−1.30 ± 0.87 C | 2.96 × 10−2 | 0.818 | |||||
| Alkanes | |||||||||||
| 2-Ethoxy-2-methyl-propane b, L2 | ↑1.01 ± 0.80 C | 3.00 × 10−2 | 0.762 | ||||||||
| Decane b, L2 | ↑0.84 ± 0.79 | 4.90 ×10−2 | 0.734 | ↑1.12 ± 0.81 E | 2.23 × 10−2 | 0.780 | |||||
| Tetradecane c, L2, * | ↑3.68 ± 1.25 E | 2.90 × 10−7 | 1.000 | ↑2.81 ± 1.07 E | 2.90 × 10−7 | 1.000 | ↑1.88 ± 0.94 E | 5.61 × 10−4 | 0.909 | ||
| Dodecane b, L2 | ↓−1.29 ± 0.87 C | 2.96 × 10−2 | 0.811 | ||||||||
| Alkenes | |||||||||||
| 2,4-Dimethyl-1-heptene b, L2 | ↑0.80 ± 0.79 C | 3.00 × 10−2 | 0.769 | ↑0.89 ± 0.78 C | 2.23 × 10−2 | 0.780 | |||||
| 3-Carene b, L1 | ↑2.32 ± 0.99 | 4.33 × 10−5 | 0.962 | ↑1.84 ± 0.90 | 2.59 × 10−4 | 0.940 | |||||
| Aldehydes | |||||||||||
| 4-Methylbenzaldehyde b, L2 | ↑2.00 ± 0.94 C | 1.60 × 10−4 | 0.936 | ↑1.69 ± 0.88 C | 1.10 × 10−3 | 0.899 | |||||
| Benzaldehyde b, L1 | ↓−1.59 ± 0.86 C | 2.74 × 10−3 | 0.863 | ||||||||
| 3-Methylbenzaldehyde c, L2 | ↑1.75 ± 0.89 E | 2.34 × 10−5 | 0.946 | ↑2.03 ± 0.93 E | 9.32 × 10−6 | 0.959 | |||||
| Acetaldehyde c, L1 | ↓−10.93 ± 3.06 C | 2.90 × 10−7 | 1.000 | ↓−4.48 ± 1.43 C | 2.90 × 10−7 | 1.000 | ↓−4.77 ± 1.55 C | 5.18 × 10−6 | 0.992 | ||
| Formaldehyde c, L2 | ↑3.34 ± 1.18 C | 2.90 × 10−7 | 1.000 | ↑2.58 ± 1.02 C | 2.90 × 10−7 | 1.000 | ↑3.13 ± 1.18 C | 5.18 × 10−6 | 1.000 | ||
| Decanal b, L1, * | ↓−1.72 ± 0.90 C | 1.61 × 10−3 | 0.872 | ↓−1.87 ± 0.90 C | 3.00 × 10−3 | 0.857 | |||||
| Benzene Derivatives | |||||||||||
| Ethylbenzene b, L2 | ↑1.46 ± 0.86 | 3.58 × 10−4 | 0.917 | ↑1.00 ± 0.79 E | 2.42 × 10−3 | 0.875 | |||||
| Styrene b, L2, * | ↑1.32 ± 0.84 E | 1.61 × 10−3 | 0.871 | ||||||||
| Xylene b, L2 | ↑1.61 ± 0.88 E | 4.33 × 10−5 | 0.962 | ↑1.11 ± 0.81 E | 6.27 × 10−4 | 0.917 | |||||
| Ketones | |||||||||||
| Acetophenone b, L2 | ↓− 0.96 ± 0.80C | 4.31 × 10−2 | 0.750 | ||||||||
| Cyclohexanone b, L2, * | ↓− 3.29 ± 1.19 C | 2.88 × 10−6 | 1.000 | ↓−1.53 ± 0.85 C | 2.74 × 10−3 | 0.863 | ↓−1.54 ± 0.91 C | 6.04 × 10−3 | 0.879 | ||
| Acetone c, L2 | ↑2.12 ± 0.94 E | 5.63 × 10−5 | 0.929 | ↑2.22 ± 0.96 E | 7.25 × 10−6 | 0.964 | ↑1.33 ± 0.86 E | 6.56 × 10−3 | 0.833 | ||
| Unknowns | |||||||||||
| Un (RT 8.40, m/z 69) b, L4 | ↑1.44 ± 0.86 C | 1.43 × 10−2 | 0.801 | ↑1.53 ± 0.85 C | 3.31 × 10−3 | 0.851 | |||||
| Un (RT 10.18, m/z 58) c, L4 | ↑3.64 ± 1.24 E | 2.90 × 10−7 | 1.000 | ↑3.07 ± 1.12 E | 2.90 × 10−7 | 1.000 | ↑2.04 ± 0.97 E | 5.18 × 10−6 | 0.992 | ||
| Un (RT 12.82. m/z 69) b, L4 | ↑0.98 ± 0.79 | 2.40 × 10−2 | 0.774 | ||||||||
| Un (RT 16.64, m/z 61) b, L4 | ↓− 4.16 ± 1.38 | 2.88 × 10−6 | 1.000 | ↓−4.39 ± 1.41 E | 3.73 × 10−6 | 1.000 | |||||
| Metabolite | Caki-2 (n = 12) vs. HK-2 (n = 12) | ACHN (n = 12) vs. HK-2 (n = 12) | |||||
|---|---|---|---|---|---|---|---|
| ES ± SE | p-Value a | AUC | ES ± SE | p-Value a | AUC | ||
| Alcohols | |||||||
| 2-Ethylhexanol b, L2, * | ↑2.51 ± 1.05 E | 2.22 × 10−5 | 0.972 | ↑4.29 ± 1.44 E | 7.40 × 10−6 | 1.000 | |
| Cyclohexanol b, L2 | ↑1.45 ± 0.87 E | 2.87 × 10−3 | 0.868 | ||||
| Alkanes | |||||||
| Tetradecane c, L2, * | ↑3.15 ± 1.18 E | 8.63 × 10−7 | 1.000 | ↑2.50 ± 1.05 E | 1.73 × 10−6 | 1.000 | |
| Aldehydes | |||||||
| 4-Methylbenzaldehyde b, L2 | ↓−1.84 ± 0.93 C | 4.78 × 10−3 | 0.875 | ||||
| 3-Methylbenzaldehyde c, L2 | ↑0.81 ± 0.81 C | 1.21 × 10−2 | 0.799 | ||||
| Acetaldehyde c, L1 | ↓−9.59 ± 2.82 C | 8.63 × 10−7 | 1.000 | ↓−10.95 ± 3.19 C | 1.73 × 10−6 | 1.000 | |
| Formaldehyde c, L2 | ↑4.39 ± 1.46C | 8.63 × 10−7 | 1.000 | ↑2.34 ± 1.02 C | 7.25 × 10−6 | 0.979 | |
| Ketones | |||||||
| 2-Pentadecanone b, L1 | ↓−2.64 ± 1.07 | 9.86 × 10−6 | 0.986 | ||||
| Acetophenone b, L2 | ↓−4.85 ± 1.57 C | 3.70 × 10−6 | 1.000 | ||||
| Cyclohexanone b, L2, * | ↓−7.09 ± 2.15 C | 3.70 × 10−6 | 1.000 | ↓−1.40 ± 0.87 C | 4.78 × 10−3 | 0.868 | |
| Acetone c, L2 | ↑4.63 ± 1.52 E | 8.63 × 10−6 | 1.000 | ↑2.89 ± 1.13 E | 1.04 × 10−5 | 0.972 | |
| Unknowns | |||||||
| Un (RT 9.80, m/z 59) b, L4 | ↑1.55 ± 0.89 C | 3.05 × 10−3 | 0.861 | ||||
| Un (RT 10.18, m/z 58) c, L4 | ↑4.65 ± 1.52 E | 8.63 × 10−7 | 1.000 | ↑3.38 ± 1.23 E | 1.73 × 10−6 | 1.000 | |
| Un (RT 12.82. m/z 69) b, L4 | ↓−0.99 ± 0.82 C | 2.47 × 10−6 | 0.785 | ||||
| Un (RT 16.64, m/z 61) b, L4 | ↓−1.23 ± 0.85 E | 1.13 × 10−2 | 0.833 | ||||
| Metabolite | Metastatic ccRCC (n = 11) vs. Non-Metastatic ccRCC (n = 27) | Metastatic pRCC (n = 12) vs. Non-Metastatic pRCC (n = 12) | ccRCC (n = 38) vs. pRCC (n = 24) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ES ± SE | p-Value a | AUC | ES ± SE | p-Value a | AUC | ES ± SE | p-Value a | AUC | |||
| Alcohols | |||||||||||
| 2-Ethylhexanol b, L2 | ↑1.20 ± 0.84 E | 1.19 × 10−2 | 0.828 | ↓−0.80 ± 0.52 E | 1.88 × 10−3 | 0.730 | |||||
| Cyclohexanol b, L2 | ↑1.83 ± 0.80 E | 1.71 × 10−4 | 0.882 | ↓−2.61 ± 1.07 | 7.40 × 10−6 | 0.993 | ↑1.15 ± 0.54 C | 2.04 × 10−5 | 0.813 | ||
| Alkanes | |||||||||||
| 2-Ethoxy-2-methyl-propane b, L2 | ↓−0.99 ± 0.72 C | 7.42 × 10−3 | 0.781 | ↑0.95 ± 0.53 | 1.13 × 10−4 | 0.763 | |||||
| Decane b, L2 | ↓−2.54 ± 0.89 C | 1.05 × 10−7 | 0.989 | ↓−0.96 ± 0.82 C | 1.67 × 10−2 | 0.806 | ↓−0.98 ± 0.53 | 2.89 × 10−4 | 0.728 | ||
| Dodecane b, L2 | ↓−2.33 ± 0.86 | 1.35 × 10−6 | 0.959 | ↓−1.28 ± 0.85 | 1.81 × 10−2 | 0.799 | ↑1.03 ± 0.54 | 9.68 × 10−5 | 0.782 | ||
| 4-Methylheptane b, L2 | ↓−1.40 ± 0.75 C | 2.60 × 10−4 | 0.872 | ↑1.28 ± 0.55 C | 1.31 × 10−6 | 0.839 | |||||
| Tetradecane c, L2 | ↓−1.64 ± 0.90 E | 5.78 × 10−4 | 0.896 | ||||||||
| Alkenes | |||||||||||
| 2,4-Dimethyl-1-heptene b, L2 | ↓−1.60 ± 0.77 C | 5.02 × 10−5 | 0.906 | ↑1.29 ± 0.55 C | 1.31 × 10−6 | 0.842 | |||||
| 3-Carene b, L1 | ↓−2.29 ± 0.86 C | 2.99 × 10−8 | 1.000 | ↑1.36 ± 0.56 | 6.99 × 10−7 | 0.850 | |||||
| Aldehydes | |||||||||||
| 4-Methylbenzaldehyde b, L2 | ↓−2.26 ±0.85 C | 1.89 × 10−7 | 0.983 | ↓−1.23 ± 0.85 C | 2.79 × 10−2 | 0.778 | ↑1.28 ± 0.55 C | 2.22 × 10−6 | 0.827 | ||
| 3-Methylbenzaldehyde c, L2 | ↓−0.97 ± 0.82 C | 3.84 × 10−4 | 0.910 | ||||||||
| Acetaldehyde c, L1 | ↓−1.90 ± 0.94 C | 3.84 × 10−4 | 0.917 | ||||||||
| Formaldehyde c, L2 | ↓−1.29 ± 0.85 C | 5.56 × 10−3 | 0.826 | ||||||||
| Decanal b, L1 | ↑2.58 ± 0.90 E | 7.25 × 10−7 | 0.969 | ↓−1.07 ± 0.83 E | 1.13 ×10−2 | 0.833 | ↑0.54 ± 0.51 E | 3.29 × 10−2 | 0.654 | ||
| Benzene Derivatives | |||||||||||
| Styrene b, L2 | ↓−1.46 ± 0.76 | 6.20 × 10−4 | 0.852 | ||||||||
| Xylene b, L2 | ↓−0.67 ± 0.70 E | 3.38 × 10−3 | 0.808 | ||||||||
| Ethylbenzene b, L2 | ↓−0.51± 0.70 C | 7.12 × 10−3 | 0.784 | ||||||||
| Ketones | |||||||||||
| 2-Pentadecanone b, L1 | ↑2.80 ± 1.11 E | 1.11 × 10−5 | 0.986 | ↑0.84 ± 0.53 E | 8.89 × 10−4 | 0.734 | |||||
| 4-Methyl-2-hexanone b, L2 | ↑0.97 ± 0.53 | 9.68 × 10−5 | 0.768 | ||||||||
| Acetone c, L2 | ↓−1.97 ± 0.95 E | 3.84 × 10−4 | 0.910 | ||||||||
| Acetophenone b, L2 | ↑5.83 ± 1.82 C | 5.55 × 10−6 | 1.000 | ↓−1.40 ± 0.56 C | 2.03 × 10−4 | 0.761 | |||||
| Cyclohexanone b, L2 | ↑8.14 ± 2.44 C | 7.40 × 10−6 | 1.000 | ||||||||
| Unknowns | |||||||||||
| Un (RT 8.40, m/z 69) b, L4 | ↓−2.02 ± 0.82 C | 2.86 × 10−6 | 0.949 | ↓−1.31 ± 0.55 E | 6.89 × 10−6 | 0.795 | |||||
| Un (RT 9.45 m/z 71) b, L4 | ↑1.79 ± 0.92 | 1.56 × 10−2 | 0.813 | ||||||||
| Un (RT 9.80, m/z 59) b, L4 | ↓−1.79 ± 0.92 | 4.33 × 10−4 | 0.924 | ||||||||
| Un (RT 10.18, m/z 58) c, L4 | ↓−1.69 ± 0.91 E | 3.84 × 10−4 | 0.917 | ||||||||
| Un (RT 12.82. m/z 69) b, L4 | ↑1.70 ± 0.79 C | 2.00 × 10−5 | 0.918 | ||||||||
| Un (RT 16.64, m/z 61) b, L4 | ↑3.58 ± 1.06 E | 8.00 × 10−7 | 0.966 | ↑1.40 ± 0.56 C | 6.99 × 10−7 | 0.844 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amaro, F.; Pinto, J.; Rocha, S.; Araújo, A.M.; Miranda-Gonçalves, V.; Jerónimo, C.; Henrique, R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach. Metabolites 2020, 10, 174. https://doi.org/10.3390/metabo10050174
Amaro F, Pinto J, Rocha S, Araújo AM, Miranda-Gonçalves V, Jerónimo C, Henrique R, Bastos MdL, Carvalho M, Guedes de Pinho P. Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach. Metabolites. 2020; 10(5):174. https://doi.org/10.3390/metabo10050174
Chicago/Turabian StyleAmaro, Filipa, Joana Pinto, Sílvia Rocha, Ana Margarida Araújo, Vera Miranda-Gonçalves, Carmen Jerónimo, Rui Henrique, Maria de Lourdes Bastos, Márcia Carvalho, and Paula Guedes de Pinho. 2020. "Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach" Metabolites 10, no. 5: 174. https://doi.org/10.3390/metabo10050174
APA StyleAmaro, F., Pinto, J., Rocha, S., Araújo, A. M., Miranda-Gonçalves, V., Jerónimo, C., Henrique, R., Bastos, M. d. L., Carvalho, M., & Guedes de Pinho, P. (2020). Volatilomics Reveals Potential Biomarkers for Identification of Renal Cell Carcinoma: An In Vitro Approach. Metabolites, 10(5), 174. https://doi.org/10.3390/metabo10050174