Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results
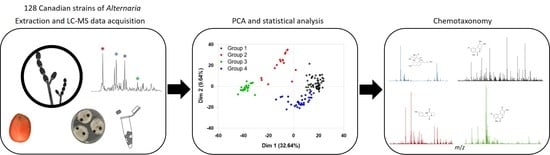
2.1. Principal Component Analysis (PCA) of Secondary Metabolites from Canadian Species of Alternaria
2.2. Statistical and Metabolomic Analysis of Detected Canadian Alternaria Populations
3. Discussion
4. Materials and Methods
4.1. Fungal Material and Identification
4.2. Agar Plug Extraction
4.3. LC-HRMS Analysis
4.4. Principal Component Analysis (PCA) and k-Means Clustering Analysis
4.5. Metabolomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patriarca, A.; Azcarate, M.P.; Terminiello, L.; Fernández Pinto, V. Mycotoxin production by Alternaria strains isolated from Argentinean wheat. Int. J. Food Microbiol. 2007, 119, 219–222. [Google Scholar] [CrossRef]
- Andersen, B.; Nielsen, K.F.; Fernández Pinto, V.; Patriarca, A. Characterization of Alternaria strains from Argentinean blueberry, tomato, walnut and wheat. Int. J. Food Microbiol. 2015, 196, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tralamazza, S.M.; Piacentini, K.C.; Iwase, C.H.T.; Rocha, L.d.O. Toxigenic Alternaria species: Impact in cereals worldwide. Curr. Opin. Food Sci. 2018, 23, 57–63. [Google Scholar] [CrossRef]
- Ayer, W.A.; Pena-Rodriguez, L.M. Metabolites produced by Alternaria brassicae, the black spot pathogen of canola. Part 1, the phytotoxic components. J. Nat. Prod. 1987, 50, 400–407. [Google Scholar] [CrossRef]
- European Food Safety Authority; Arcella, D.; Eskola, M.; Ruiz, J.A.G. Dietary exposure assessment to Alternaria toxins in the European population. EFSA J. 2016, 14. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific opinion on the risks for animal and public health related to the presence of Alternaria toxins in feed and food. EFSA J. 2011, 9, 97. [Google Scholar]
- Harimoto, Y.; Tsuge, T.; Akimitsu, K.; Ohtani, K.; Otani, H.; Egusa, M.; Kodama, M.; Akagi, Y.; Yamamoto, M. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar]
- Takaoka, S.; Kurata, M.; Harimoto, Y.; Hatta, R.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. Complex regulation of secondary metabolism controlling pathogenicity in the phytopathogenic fungus Alternaria alternata. New Phytol. 2014, 202, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.J.; Cherry, W.; Soper, K.; Langlois, I.; Vijay, H.M. Examination of Alternaria alternata mutagenicity and effects of nitrosylation using the ames salmonella test. Teratog. Carcinog. Mutagenesis 2001, 21, 261–274. [Google Scholar] [CrossRef]
- Brugger, E.-M.; Wagner, J.; Schumacher, D.M.; Koch, K.; Podlech, J.; Metzler, M.; Lehmann, L. Mutagenicity of the mycotoxin alternariol in cultured mammalian cells. Toxicol. Lett. 2006, 164, 221–230. [Google Scholar] [CrossRef]
- Scott, P.M.; Zhao, W.; Feng, S.; Lau, B.P.-Y. Alternaria toxins alternariol and alternariol monomethyl ether in grain foods in Canada. Mycotoxin Res. 2012, 28, 261–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Perre, E.; Deschuyffeleer, N.; Jacxsens, L.; Vekeman, F.; Van Der Hauwaert, W.; Asam, S.; Rychlik, M.; Devlieghere, F.; De Meulenaer, B. Screening of moulds and mycotoxins in tomatoes, bell peppers, onions, soft red fruits and derived tomato products. Food Control. 2014, 37, 165–170. [Google Scholar] [CrossRef]
- Lau, B.P.Y.; Scott, P.M.; Lewis, D.A.; Kanhere, S.R.; Cléroux, C.; Roscoe, V.A. Liquid chromatography–mass spectrometry and liquid chromatography–tandem mass spectrometry of the Alternaria mycotoxins alternariol and alternariol monomethyl ether in fruit juices and beverages. J. Chromatogr. A 2003, 998, 119–131. [Google Scholar] [CrossRef]
- Asam, S.; Rychlik, M. Potential health hazards due to the occurrence of the mycotoxin tenuazonic acid in infant food. Eur. Food Res. Technol. 2013, 236, 491–497. [Google Scholar] [CrossRef]
- Rychlik, M.; Lepper, H.; Weidner, C.; Asam, S. Risk evaluation of the Alternaria mycotoxin tenuazonic acid in foods for adults and infants and subsequent risk management. Food Control. 2016, 68, 181–185. [Google Scholar] [CrossRef]
- Renaud, J.B.; Kelman, M.J.; Qi, T.F.; Seifert, K.A.; Sumarah, M.W. Product ion filtering with rapid polarity switching for the detection of all fumonisins and AAL-toxins. Rapid Commun. Mass Spectrom. 2015, 29, 2131–2139. [Google Scholar] [CrossRef]
- Shier, W.T.; Abbas, H.K.; Mirocha, C.J. Toxicity of the mycotoxins Fumonisins B1 and B2 and Alternaria alternata f. sp. lycopersici toxin (AAL) in cultured mammalian cells. Mycopathologia 1991, 116, 97–104. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene; IARC monographs on the evaluation of carcinogenic risks to humans; IARC: Lyon, France, 2002. [Google Scholar]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [Green Version]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, D.P.; Rotondo, F.; Gannibal, P.B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 2015, 15, 3. [Google Scholar] [CrossRef]
- Hatta, R.; Ito, K.; Hosaki, Y.; Tanaka, T.; Tanaka, A.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 2002, 161, 59–70. [Google Scholar]
- Masunaka, A.; Ohtani, K.; Peever, T.L.; Timmer, L.W.; Tsuge, T.; Yamamoto, M.; Yamamoto, H.; Akimitsu, K. An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT- and ACR-toxins. Phytopathology 2005, 95, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Akagi, Y.; Taga, M.; Yamamoto, M.; Tsuge, T.; Fukumasa-Nakai, Y.; Otani, H.; Kodama, M. Chromosome constitution of hybrid strains constructed by protoplast fusion between the tomato and strawberry pathotypes of Alternaria alternata. J. Gen. Plant Pathol. 2009, 75, 101. [Google Scholar] [CrossRef]
- Salamiah; Akamatsu, H.; Fukumasa-Nakai, Y.; Otani, H.; Kodama, M. Construction and genetic analysis of hybrid strains between apple and tomato pathotypes of Alternaria alternata by protoplast fusion. J. Gen. Plant Pathol. 2001, 67, 97–105. [Google Scholar] [CrossRef]
- Salamiah; Fukumasa-Nakai, Y.; Akamatsu, H.; Otani, H.; Kohmoto, K.; Kodama, M. Genetic analysis of pathogenicity and host-specific toxin production of Alternaria alternata tomato pathotype by protoplast fusion. J. Gen. Plant Pathol. 2001, 67, 7–14. [Google Scholar]
- da Cruz Cabral, L.; Rodriguero, M.; Stenglein, S.; Fog Nielsen, K.; Patriarca, A. Characterization of small-spored Alternaria from Argentinean crops through a polyphasic approach. Int. J. Food Microbiol. 2017, 257, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Patriarca, A.; da Cruz Cabral, L.; Pavicich, M.A.; Nielsen, K.F.; Andersen, B. Secondary metabolite profiles of small-spored Alternaria support the new phylogenetic organization of the genus. Int. J. Food Microbiol. 2019, 291, 135–143. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef]
- Andersen, B.; Sørensen, J.L.; Nielsen, K.F.; van den Ende, B.G.; de Hoog, S. A polyphasic approach to the taxonomy of the Alternaria infectoria species–group. Fungal Genet. Biol. 2009, 46, 642–656. [Google Scholar] [CrossRef]
- Andersen, B.; Thrane, U. Differentiation of Alternaria infectoria and Alternaria alternata based on morphology, metabolite profiles, and cultural characteristics. Can. J. Microbiol. 1996, 42, 685–689. [Google Scholar] [CrossRef]
- Andersen, B.; Krøger, E.; Roberts, R.G. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol. Res. 2001, 105, 291–299. [Google Scholar] [CrossRef]
- Andrew, M.; Peever, T.L.; Pryor, B. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 2009, 101, 95–109. [Google Scholar] [CrossRef] [Green Version]
- Andersen, B.; Dongo, A.; Pryor, B.M. Secondary metabolite profiling of Alternaria dauci, A. porri, A. solani, and A. Tomatophila. Mycol. Res. 2008, 112, 241–250. [Google Scholar] [CrossRef]
- Somma, S.; Amatulli, M.T.; Masiello, M.; Moretti, A.; Logrieco, A.F. Alternaria species associated to wheat black point identified through a multilocus sequence approach. Int. J. Food Microbiol. 2019, 293, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.W.; Skolko, A.J. Notes on seed-borne fungi: Ii. Alternaria. Can. J. Res. 1944, 22c, 217–234. [Google Scholar] [CrossRef]
- Pryor, B.M.; Bigelow, D.M. Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia 2003, 95, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.-S.; Motoyama, T.; Osada, H. Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS–PKS hybrid enzyme. Nat. Commun. 2015, 6, 8758. [Google Scholar] [CrossRef] [Green Version]
- Saha, D.; Fetzner, R.; Burkhardt, B.; Podlech, J.; Metzler, M.; Dang, H.; Lawrence, C.; Fischer, R. Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata. PLoS ONE 2012, 7, e40564. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Smedsgaard, J.; Samson, R.A.; Larsen, T.O.; Thrane, U. Fumonisin B2 production by Aspergillus niger. J. Agric. Food Chem. 2007, 55, 9727–9732. [Google Scholar] [CrossRef]
- Kelman, M.J.; Renaud, J.B.; Seifert, K.A.; Mack, J.; Sivagnanam, K.; Yeung, K.K.-C.; Sumarah, M.W. Identification of six new Alternaria sulfoconjugated metabolites by high-resolution neutral loss filtering. Rapid Commun. Mass Spectrom. 2015, 29, 1805–1810. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. Xcms: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Böttcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [Green Version]
- Want, E.J.; Benton, H.P.; Ebbels, T.M.D. Correction of Mass Calibration Gaps in Liquid Chromatography–Mass Spectrometry Metabolomics Data. Bioinformatics 2010, 26, 2488–2489. [Google Scholar]
- McMillan, A.; Rulisa, S.; Sumarah, M.; Macklaim, J.M.; Renaud, J.; Bisanz, J.E.; Gloor, G.B.; Reid, G. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci. Rep. 2015, 5, 14174. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations, Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]


| Metabolite | Abbr. | RT (min) | Populations of Canadian Strains (% of Strains) | |||
|---|---|---|---|---|---|---|
| Group 1 (n = 57) | Group 2 (n = 16) | Group 3 (n = 25) | Group 4 (n = 30) | |||
| infectopyrone | IPy | 3.03 | - | - | 100 | - |
| phomapyrone A | PPyA | 3.73 | - | - | 100 | - |
| phomapyrone B | PPyB | 2.83 | - | - | 85 | - |
| phomapyrone D | PPyD | 3.04 | - | - | 100 | - |
| phomapyrone E/G | PPyE/G | 2.99 | - | - | 100 | - |
| phomapyrone F | PPyF | 3.01 | - | - | 50 | - |
| tenuazonic acid | TeA | 3.05 | 100 | - | - | 100 |
| iso-tenuazonic acid derivative | IsoTeA | 2.81 | 100 | - | - | 100 |
| alternariol * | AOH | 3.1 | 100 | 94 | - | 91 |
| alternariol monomethyl ether * | AME | 3.53 | 100 | 100 | - | 94 |
| altenusin * | ALU | 2.97 | 100 | 71 | - | 55 |
| desmethylaltenusin * | DMA | 2.75 | 23 | 18 | - | - |
| dehydroaltenusin * | DHA | 3.26 | 72 | 41 | - | 9 |
| alternarienoic acid * | AlA | 2.71 | 100 | 71 | - | 30 |
| altenuene | ALT | 2.87 | 70 | 53 | - | - |
| altechromone A | ALCA | 2.82 | 79 | 53 | - | 30 |
| altechromone B | ALCB | 2.72 | 98 | 82 | - | 12 |
| altertoxin I * | ALTX-I | 2.53 | 33 | 59 | - | 36 |
| altertoxin II * | ALTX-II | 2.93 | 16 | 35 | - | 30 |
| altertoxin III * | ALTX-III | 3.27 | 26 | 18 | - | 9 |
| tentoxin | TTX | 3.11 | 30 | 29 | - | 21 |
| dihydrotentoxin | D-TTX | 3.13 | 33 | 24 | - | 15 |
| altersetin * | ALS | 4.37 | 68 | 53 | - | 67 |
| pyrenochaetic acid A | PyrA | 2.63 | 93 | 71 | - | 21 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelman, M.J.; Renaud, J.B.; Seifert, K.A.; Mack, J.; Yeung, K.K.-C.; Sumarah, M.W. Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry. Metabolites 2020, 10, 238. https://doi.org/10.3390/metabo10060238
Kelman MJ, Renaud JB, Seifert KA, Mack J, Yeung KK-C, Sumarah MW. Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry. Metabolites. 2020; 10(6):238. https://doi.org/10.3390/metabo10060238
Chicago/Turabian StyleKelman, Megan J., Justin B. Renaud, Keith A. Seifert, Jonathan Mack, Ken K.-C. Yeung, and Mark W. Sumarah. 2020. "Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry" Metabolites 10, no. 6: 238. https://doi.org/10.3390/metabo10060238
APA StyleKelman, M. J., Renaud, J. B., Seifert, K. A., Mack, J., Yeung, K. K. -C., & Sumarah, M. W. (2020). Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry. Metabolites, 10(6), 238. https://doi.org/10.3390/metabo10060238