Abiotic and Biotic Damage of Microalgae Generate Different Volatile Organic Compounds (VOCs) for Early Diagnosis of Algal Cultures for Biofuel Production
Abstract
:1. Introduction
2. Results
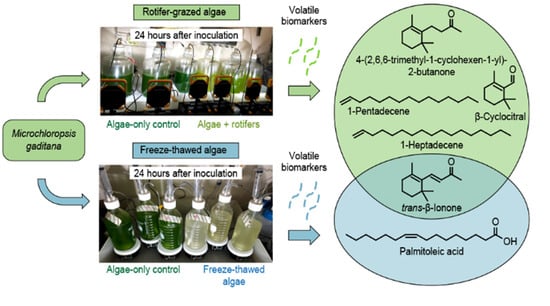
2.1. Monitoring Algal Concentration during Rotifer-Grazing and Freeze–Thaw Experiments
2.1.1. Rotifer-Grazing Experiments
2.1.2. Freeze–Thaw Experiments
2.2. Monitoring VOC Emissions during Rotifer-Grazing and Freeze–Thaw Experiments
2.2.1. Rotifer-Grazing Experiments
2.2.2. Freeze–Thaw Experiments
2.2.3. Comparison of Algae Crop Damage Mechanisms
3. Discussion
4. Materials and Methods
4.1. Microalgae and Rotifer Cultures
4.2. Freeze–Thaw Algal Experiments
4.3. Daily Timepoints to Monitor Viability of Algal Cultures for A + R and FTA Experiments
4.4. SPME Headspace Sampling and GC-MS Data Acquisition
4.5. Volatilomics Analysis and Requirements for Defining a ‘Biomarker’ for This Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Energy Agency (IEA). Global Energy Review 2020; summarized under “Energy Consumption by the Numbers”; IEA: Paris, France, 2020. Available online: https://www.capp.ca/energy/world-energy-needs/ (accessed on 15 September 2021).
- Ullah, K.; Ahmad, M.; Sofia; Sharma, V.K.; Lu, P.; Harvey, A.; Zafar, M.; Sultana, S. Assessing the Potential of Algal Biomass Opportunities for Bioenergy Industry: A Review. Fuel 2015, 143, 414–423. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft Genome Sequence and Genetic Transformation of the Oleaginous Alga Nannochloropsis Gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, C.L.; Lane, T.W. Operational, Prophylactic, and Interdictive Technologies for Algal Crop Protection. In Grand Challenges in Algae Biotechnology; Hallmann, A., Rampelotto, P.H., Eds.; Springer International Publishing: Cham, Denmark, 2019; pp. 35–70. [Google Scholar]
- Fawley, M.W.; Jameson, I.; Fawley, K.P. The Phylogeny of the Genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with Descriptions of N. Australis sp. nov. and Microchloropsis gen. nov. Phycologia 2015, 54, 545–552. [Google Scholar] [CrossRef]
- Doan, T.T.Y.; Sivaloganathan, B.; Obbard, J.P. Screening of Marine Microalgae for Biodiesel Feedstock. Biomass Bioenergy 2011, 35, 2534–2544. [Google Scholar] [CrossRef]
- Jinkerson, R.E.; Radakovits, R.; Posewitz, M.C. Genomic Insights from the Oleaginous Model Alga Nannochloropsis Gaditana. Bioengineered 2013, 4, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Cancela, A.; Pérez, L.; Febrero, A.; Sánchez, A.; Salgueiro, J.L.; Ortiz, L. Exploitation of Nannochloropsis Gaditana Biomass for Biodiesel and Pellet Production. Renew. Energy 2019, 133, 725–730. [Google Scholar] [CrossRef]
- Taher, H.; Giwa, A.; Abusabiekeh, H.; Al-Zuhair, S. Biodiesel Production from Nannochloropsis Gaditana Using Supercritical CO2 for Lipid Extraction and Immobilized Lipase Transesterification: Economic and Environmental Impact Assessments. Fuel Process. Technol. 2020, 198, 106249. [Google Scholar] [CrossRef]
- López, E.N.; Medina, A.R.; Moreno, P.A.G.; Cerdán, L.E.; Valverde, L.M.; Grima, E.M. Biodiesel Production from Nannochloropsis Gaditana Lipids through Transesterification Catalyzed by Rhizopus oryzae Lipase. Bioresour. Technol. 2016, 203, 236–244. [Google Scholar] [CrossRef]
- Lavens, P.; Sorgeloos, P. Manual on the Production and Use of Live Food for Aquaculture; FAO Fisheries Technical Paper. No. 361; FAO: Rome, Italy, 1996; 295p. [Google Scholar]
- Bartosh, Y.; Banks, C. Algal growth response and survival in a range of light and temperature conditions: Implications for non-steady-state conditions in waste stabilisation ponds. Water Sci. Technol. 2007, 55, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Carney, L.T.; Lane, T.W. Parasites in Algae Mass Culture. Front. Microbiol. 2014, 5, 278. [Google Scholar] [CrossRef] [Green Version]
- Harmon, V.L.; Wolfrum, E.; Knoshaug, E.P.; Davis, R.; Laurens, L.M.; Pienkos, P.T.; McGowen, J. Reliability metrics and their management implications for open pond algae cultivation. Algal Res. 2021, 55, 102249. [Google Scholar] [CrossRef]
- McBride, R.C.; Lopez, S.; Meenach, C.; Burnett, M.; Lee, P.A.; Nohilly, F.; Behnke, C. Contamination Management in Low Cost Open Algae Ponds for Biofuels Production. Ind. Biotechnol. 2014, 10, 221–227. [Google Scholar] [CrossRef]
- Borowitzka, M.A. Culturing Microalgae in Outdoor Ponds. In Algal Culturing Techniques; Anderson, R.A., Ed.; Elsiver Ac-ademic Press: London, UK, 2005; pp. 205–218. [Google Scholar]
- Wang, Y.; Castillo-Keller, M.; Eustance, E.; Sommerfeld, M. Early Detection and Quantification of Zooplankton Grazers in Algal Cultures by Flowcam. Algal Res. 2017, 21, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Day, J.G.; Thomas, N.J.; Achilles-Day, U.E.M.; Leakey, R.J.G. Early Detection of Protozoan Grazers in Algal Biofuel Cultures. Bioresour. Technol. 2012, 114, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Carney, L.T.; McBride, R.C.; Smith, V.H.; Lane, T.W. Molecular Diagnostic Solutions in Algal Cultivation Systems. In Micro-Algal Production for Biomass and High Value Products; Slocombe, S.P., Benemann, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 2016; p. 22. [Google Scholar]
- Bos, L.D.; Sterk, P.J.; Schultz, M.J. Volatile Metabolites of Pathogens: A Systematic Review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasu, M.; Touhara, K. The Scent of Disease: Volatile Organic Compounds of the Human Body Related to Disease and Disorder. J. Biochem. 2011, 150, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Achyuthan, K.E.; Harper, J.C.; Manginell, R.P.; Moorman, M.W. Volatile Metabolites Emission by In Vivo Microalgae-an Overlooked Opportunity? Metabolites 2017, 7, 39. [Google Scholar] [CrossRef]
- Hosoglu, M.I.; Karagul-Yuceer, Y.; Guneser, O. Aroma Characterization of Heterotrophic Microalgae Crypthecodinium cohnii Using Solid-Phase Microextraction and Gas Chromatography–Mass Spectrometry/Olfactometry During Different Growth Phases. Algal. Res. 2020, 49, 101928. [Google Scholar] [CrossRef]
- Reese, K.L.; Fisher, C.L.; Lane, P.D.; Jaryenneh, J.D.; Moorman, M.W.; Jones, A.D.; Frank, M.; Lane, T.W. Chemical Profiling of Volatile Organic Compounds in the Headspace of Algal Cultures as Early Biomarkers of Algal Pond Crashes. Sci. Rep. 2019, 9, 13866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, C.L.; Lane, P.D.; Russell, M.; Maddalena, R.; Lane, T.W. Low Molecular Weight Volatile Organic Compounds Indicate Grazing by the Marine Rotifer Brachionus plicatilis on the Microalgae Microchloropsis Salina. Metabolites 2020, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Regmi, B.P.; Agah, M. Micro Gas Chromatography: An Overview of Critical Components and Their Integration. Anal. Chem. 2018, 90, 13133–13150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirayama, K.; Ogawa, S. Fundamental Studies on Physiology of Rotifer for Its Mass Culture-I Filter Feeding of Rotifer. Nippon. Suisan. Gakkaishi. 1972, 38, 1207–1214. [Google Scholar] [CrossRef] [Green Version]
- Gregor, J.; Maršálek, B. Freshwater Phytoplankton Quantification by Chlorophyll A: A Comparative Study of In Vitro, In Vivo and In Situ Methods. Water Res. 2004, 38, 517–522. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, W.; Xu, X.; Zhang, H.; Wang, J.; Xian, M. Production of Free Monounsaturated Fatty Acids by Metabolically Engineered Escherichia coli. Biotechnol. Biofuels 2014, 7, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudahettige, N.P.; Pickova, J.; Gentili, F.G. Stressing Algae for Biofuel Production: Biomass and Biochemical Composition of Scenedesmus dimorphus and Selenastrum minutum Grown in Municipal Untreated Wastewater. Front. Energy Res. 2018, 6, 132. [Google Scholar] [CrossRef]
- El-Kassas, H.Y. Growth and Fatty Acid Profile of the Marine Microalga Picochlorum Sp. Grown under Nutrient Stress Conditions. Egypt. J. Aquat. Res. 2013, 39, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.; Cortina-Burgueno, A.; Freire, I.; Otero, A. Effect of nutritional status and concentration of Nannochloropsis gaditana as enrichment diet for the marine rotifer Brachionus sp. Aquaculture 2018, 491, 351–357. [Google Scholar] [CrossRef]
- Robertson, R.C.; Guihéneuf, F.; Bahar, B.; Schmid, M.; Stengel, D.B.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. The Anti-Inflammatory Effect of Algae-Derived Lipid Extracts on Lipopolysaccharide (Lps)-Stimulated Human Thp-1 Macrophages. Mar. Drugs 2015, 13, 5402–5424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benevides, C.M.D.J.; Veloso, M.C.D.C.; Pereira, P.A.D.P.; Andrade, J.B.D. A Chemical Study of Β-Carotene Oxidation by Ozone in an Organic Model System and the Identification of the Resulting Products. Food Chem. 2011, 126, 927–934. [Google Scholar] [CrossRef] [Green Version]
- Christaki, E.; Bonos, E.; Giannenas, I.; Florou-Paneri, P. Functional Properties of Carotenoids Originating from Algae. J. Sci. Food Agric. 2013, 93, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Carotenoid Oxidation Products as Stress Signals in Plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Β-Cyclocitral and Derivatives: Emerging Molecular Signals Serving Multiple Biological Functions. Plant Physiol. Biochem. 2020, 155, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z. Why Algae Release Volatile Organic Compounds-the Emission and Roles. Front. Microbiol. 2019, 10, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novoveská, L.; Ross, M.E.; Stanley, M.S.; Pradelles, R.; Wasiolek, V.; Sassi, J.F. Microalgal Carotenoids: A Review of Production, Current Markets, Regulations, and Future Direction. Mar. Drugs 2019, 17, 640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berges, J.A.; Franklin, D.J.; Harrison, P.J. Evolution of an Artificial Seawater Medium: Improvements in Enriched Seawater, Artificial Water over the Last Two Decades. J. Phycol. 2001, 37, 1138–1145. [Google Scholar] [CrossRef]




| Compound | Base Peak m/z | Tentative Compound Class | NIST14 ID | NIST% Match | Experimental RI | Theoretical RI | A+R Expt 1 | A+R Expt 2 | A+R Expt 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 h | 72 h | 96 h | 48 h | 72 h | 96 h | 72 h | 96 h | 120 h | |||||||
| R1 | 137 | Ketone | β-cyclocitral | 83 | 1208 | 1220 | X | X | X | X | X | X | X | X | |
| R2 | 148 | unknown | 1402 | X | X | ||||||||||
| R3 | 121 | Ketone | 4-(2,6,6-trimethyl-1-cyclohexen-1-yl)-2-butanone | 75 | 1443 | 1433 | X | X | X | ||||||
| R4 | 177 | Ketone | β-ionone | 80 | 1494 | 1486 | X | X | X | X | X | X | X | X | X |
| R5 | 57 | Alkene | 1-pentadecene | 71 | 1499 | 1492 | X | X | X | X | |||||
| R6 | 83 | Alkene | 1-heptadecene | 78 | 1662 | 1687 | X | X | X | X | X | X | X | X | |
| Compound | Base Peak m/z | Tentative Compound Class | NIST14 ID | NIST% Match | Experimental RI | Theoretical RI | FTA Expt 1 | FTA Expt 2 | FTA Expt 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 1 h | 6 h | 24 h | 48 h | 1 h | 6 h | 24 h | 48 h | |||||||
| F1 | 193 | 834 | X | X | ||||||||||||
| F2 | 119 | 1177 | X | X | X | X | X | X | ||||||||
| F3 | 57 | 1265 | X | X | X | X | X | |||||||||
| F4 | 79 | 1288 | X | X | X | |||||||||||
| F5 | 81 | 1294 | X | X | X | |||||||||||
| F6 | 85 | 1381 | X | X | X | |||||||||||
| F7 | 177 | Ketone | β-ionone | 93 | 1495 | 1486 | X | X | X | X | X | X | X | |||
| F8 | 123 | 1498 | X | X | X | X | X | X | X | X | ||||||
| F9 | 55 | Carboxylic acid | Palmitoleic acid | 80 | 1954 | 1951 | X | X | X | |||||||
| F10 | 149 | 2821 | X | X | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reese, K.L.; Fisher, C.L.; Lane, P.D.; Jaryenneh, J.D.; Jones, A.D.; Frank, M.; Lane, T.W. Abiotic and Biotic Damage of Microalgae Generate Different Volatile Organic Compounds (VOCs) for Early Diagnosis of Algal Cultures for Biofuel Production. Metabolites 2021, 11, 707. https://doi.org/10.3390/metabo11100707
Reese KL, Fisher CL, Lane PD, Jaryenneh JD, Jones AD, Frank M, Lane TW. Abiotic and Biotic Damage of Microalgae Generate Different Volatile Organic Compounds (VOCs) for Early Diagnosis of Algal Cultures for Biofuel Production. Metabolites. 2021; 11(10):707. https://doi.org/10.3390/metabo11100707
Chicago/Turabian StyleReese, Kristen L., Carolyn L. Fisher, Pamela D. Lane, James D. Jaryenneh, A. Daniel Jones, Matthias Frank, and Todd W. Lane. 2021. "Abiotic and Biotic Damage of Microalgae Generate Different Volatile Organic Compounds (VOCs) for Early Diagnosis of Algal Cultures for Biofuel Production" Metabolites 11, no. 10: 707. https://doi.org/10.3390/metabo11100707
APA StyleReese, K. L., Fisher, C. L., Lane, P. D., Jaryenneh, J. D., Jones, A. D., Frank, M., & Lane, T. W. (2021). Abiotic and Biotic Damage of Microalgae Generate Different Volatile Organic Compounds (VOCs) for Early Diagnosis of Algal Cultures for Biofuel Production. Metabolites, 11(10), 707. https://doi.org/10.3390/metabo11100707