Untargeted Microbial Exometabolomics and Metabolomics Analysis of Helicobacter pylori J99 and jhp0106 Mutant
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the Extraction Solvent Volume for the Culture Media and Bacterial Cell Pellets
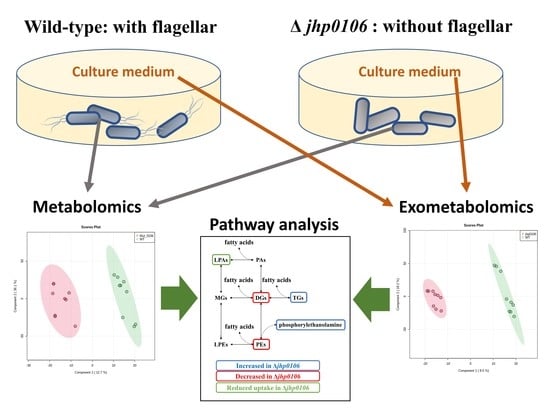
2.2. Untargeted Exometabolomic Profiling of Culture Media from H. pylori
2.3. Untargeted Metabolomic Profiling of H. pylori Pellets
2.4. Application of Untargeted Exometabolomic and Metabolomic Profiling to Exploring Flagellar Formation–Associated Metabolites and Metabolic Pathways
3. Materials and Methods
3.1. Chemicals and Materials
3.2. UHLPC-ESI-Q-TOF-MS System
3.3. Bacterial Cultivation
3.4. Sample Preparation: Culture Media and Bacterial Cell Pellets
3.5. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, J.F.; Varghese, R.S.; Zhou, B.; Nezami Ranjbar, M.R.; Zhao, Y.; Tsai, T.H.; Di Poto, C.; Wang, J.; Goerlitz, D.; Luo, Y.; et al. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J. Proteome Res. 2012, 11, 5914–5923. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.P.; Northen, T.R. Exometabolomics and MSI: Deconstructing how cells interact to transform their small molecule environment. Curr. Opin. Biotechnol. 2015, 34, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Nakahigashi, K.; Baba, T.; Robert, M.; Soga, T.; Kanai, A.; Hirasawa, T.; Naba, M.; Hirai, K.; Hoque, A.; et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 2007, 316, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 2010, 6, 364. [Google Scholar] [CrossRef]
- Depke, T.; Thoming, J.G.; Kordes, A.; Haussler, S.; Bronstrup, M. Untargeted LC-MS Metabolomics Differentiates Between Virulent and Avirulent Clinical Strains of Pseudomonas aeruginosa. Biomolecules 2020, 10, 1041. [Google Scholar] [CrossRef]
- Baidoo, E.E.K. Microbial Metabolomics: A General Overview. Methods Mol. Biol. 2019, 1859, 1–8. [Google Scholar] [CrossRef]
- Baidoo, E.E.K.; Teixeira Benites, V. Mass Spectrometry-Based Microbial Metabolomics: Techniques, Analysis, and Applications. Methods Mol. Biol. 2019, 1859, 11–69. [Google Scholar] [CrossRef]
- de Koning, W.; van Dam, K. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 1992, 204, 118–123. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas-Boas, S.G.; Aggio, R. Analysis of Intracellular Metabolites from Microorganisms: Quenching and Extraction Protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolten, C.J.; Kiefer, P.; Letisse, F.; Portais, J.C.; Wittmann, C. Sampling for metabolome analysis of microorganisms. Anal. Chem. 2007, 79, 3843–3849. [Google Scholar] [CrossRef] [PubMed]
- Canelas, A.B.; Ras, C.; Ten Pierick, A.; van Dam, J.C.; Heijnen, J.J.; van Gulik, W.M. Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics 2008, 4, 226–239. [Google Scholar] [CrossRef] [Green Version]
- Maharjan, R.P.; Ferenci, T. Global metabolite analysis: The influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 2003, 313, 145–154. [Google Scholar] [CrossRef]
- Blaser, M.J.; Atherton, J.C. Helicobacter pylori persistence: Biology and disease. J. Clin. Investig. 2004, 113, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Cover, T.L.; Blaser, M.J. Helicobacter pylori in health and disease. Gastroenterology 2009, 136, 1863–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.Y.; Kao, C.Y.; Su, M.S.; Wang, S.; Chen, Y.L.; Hu, S.T.; Chen, J.W.; Teng, C.H.; Tsai, P.J.; Wu, J.J. Glycosyltransferase Jhp0106 (PseE) contributes to flagellin maturation in Helicobacter pylori. Helicobacter 2021, 26, e12787. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, S.; Nishiumi, S.; Tagawa, R.; Yoshida, M. Alterations in metabolic pathways in gastric epithelial cells infected with Helicobacter pylori. Microb. Pathog. 2018, 124, 122–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, E.H.J.; Ng, C.G.; Goh, K.L.; Vadivelu, J.; Ho, B.; Loke, M.F. Metabolomic analysis of low and high biofilm-forming Helicobacter pylori strains. Sci. Rep. 2018, 8, 1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, C.Y.; Chen, J.W.; Wang, S.; Sheu, B.S.; Wu, J.J. The Helicobacter pylori J99 jhp0106 Gene, under the Control of the CsrA/RpoN Regulatory System, Modulates Flagella Formation and Motility. Front. Microbiol. 2017, 8, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimomura, H.; Hosoda, K.; Hayashi, S.; Yokota, K.; Hirai, Y. Phosphatidylethanolamine of Helicobacter pylori functions as a steroid-binding lipid in the assimilation of free cholesterol and 3beta-hydroxl steroids into the bacterial cell membrane. J. Bacteriol. 2012, 194, 2658–2667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.R.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, C.-Y.; Kuo, P.-Y.; Liao, H.-W. Untargeted Microbial Exometabolomics and Metabolomics Analysis of Helicobacter pylori J99 and jhp0106 Mutant. Metabolites 2021, 11, 808. https://doi.org/10.3390/metabo11120808
Kao C-Y, Kuo P-Y, Liao H-W. Untargeted Microbial Exometabolomics and Metabolomics Analysis of Helicobacter pylori J99 and jhp0106 Mutant. Metabolites. 2021; 11(12):808. https://doi.org/10.3390/metabo11120808
Chicago/Turabian StyleKao, Cheng-Yen, Pei-Yun Kuo, and Hsiao-Wei Liao. 2021. "Untargeted Microbial Exometabolomics and Metabolomics Analysis of Helicobacter pylori J99 and jhp0106 Mutant" Metabolites 11, no. 12: 808. https://doi.org/10.3390/metabo11120808
APA StyleKao, C. -Y., Kuo, P. -Y., & Liao, H. -W. (2021). Untargeted Microbial Exometabolomics and Metabolomics Analysis of Helicobacter pylori J99 and jhp0106 Mutant. Metabolites, 11(12), 808. https://doi.org/10.3390/metabo11120808