Plasma Metabolomics in a Nonhuman Primate Model of Abdominal Radiation Exposure
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sex-Specific Patterns in Response to Abdominal Irradiation Were Observed in the Plasma Metabolomics Data
2.2. Radiation Dose-Dependent and Time-Dependent Metabolic Alterations in the Plasma Metabolomics Data Were Observed
2.3. Global Metabolomic Profiles Identify Radiation Dose-Dependent and Time-Dependent Biochemical Signatures
2.4. Metabolite Alterations in Plasma Indicate Radiation-Induced GI Injury
3. Materials and Methods
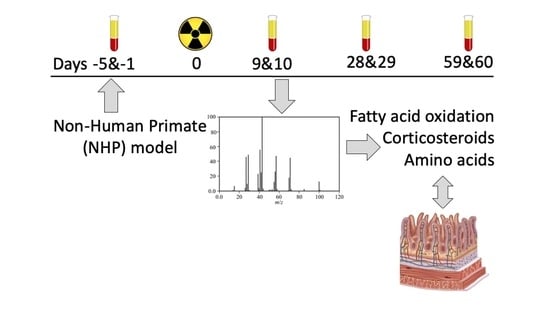
3.1. NHP System
3.2. Plasma Sample Preparation and Instrumentation
3.3. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waselenko, J.K.; MacVittie, T.J.; Blakely, W.F.; Pesik, N.; Wiley, A.L.; Dickerson, W.E.; Tsu, H.; Confer, D.L.; Coleman, C.N.; Seed, T.; et al. Medical Management of the Acute Radiation Syndrome: Recommendations of the Strategic National Stockpile Radiation Working Group. Ann. Intern. Med. 2004, 140, 1037–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dörr, H.; Meineke, V. Acute radiation syndrome caused by accidental radiation exposure-therapeutic principles. BMC Med. 2011, 9, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Olabisi, A.O. Nonhuman primates as models for the discovery and development of radiation countermeasures. Expert Opin. Drug Discov. 2017, 12, 695–709. [Google Scholar] [CrossRef]
- Singh, V.; Newman, V.L.; Berg, A.N.; MacVittie, T.J. Animal models for acute radiation syndrome drug discovery. Expert Opin. Drug Discov. 2015, 10, 497–517. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Garg, S.; Landes, R.D.; Liu, L.; Fu, Q.; Seng, J.; Boerma, M.; Thrall, K.; Hauer-Jensen, M.; Pathak, R. Differential Recovery of Small Intestinal Segments after Partial-Body Irradiation in Non-Human Primates. Radiat. Res. 2021, 196, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Fève, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 2017, 12, e0173615. [Google Scholar] [CrossRef] [Green Version]
- Moffett, J.R.; Arun, P.; Puthillathu, N.; Vengilote, R.; Ives, J.A.; Badawy, A.A.-B.; Namboodiri, A.M. Quinolinate as a Marker for Kynurenine Metabolite Formation and the Unresolved Question of NAD+ Synthesis During Inflammation and Infection. Front. Immunol. 2020, 11, 31. [Google Scholar] [CrossRef] [Green Version]
- Raison, C.L.; Dantzer, R.; Kelley, K.W.; Lawson, M.A.; Woolwine, B.; Vogt, G.; Spivey, J.R.; Saito, K.; Miller, A.H. CSF concentrations of brain tryptophan and kynurenines during immune stimulation with IFN-α: Relationship to CNS immune responses and depression. Mol. Psychiatry 2009, 15, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, K.; Hosomi, K.; Sawane, K.; Kunisawa, J. Metabolism of Dietary and Microbial Vitamin B Family in the Regulation of Host Immunity. Front. Nutr. 2019, 6, 48. [Google Scholar] [CrossRef] [Green Version]
- Oei, S.L.; Ziegler, M. ATP for the DNA Ligation Step in Base Excision Repair Is Generated from Poly(ADP-ribose). J. Biol. Chem. 2000, 275, 23234–23239. [Google Scholar] [CrossRef] [Green Version]
- Murata, M.M.; Kong, X.; Moncada, E.; Chen, Y.; Imamura, H.; Wang, P.; Berns, M.; Yokomori, K.; Digman, M.A. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol. Biol. Cell 2019, 30, 2584–2597. [Google Scholar] [CrossRef] [PubMed]
- Slominska, E.; Smolenski, R.; Szolkiewicz, M.; Leaver, N.; Rutkowski, B.; Simmonds, H.A.; Swierczynski, J. Accumulation of plasma N-methyl-2-pyridone-5-carboxamide in patients with chronic renal failure. Mol. Cell. Biochem. 2002, 231, 83–88. [Google Scholar] [CrossRef]
- Karl, J.P.; Margolis, L.; Madslien, E.H.; Murphy, N.E.; Castellani, J.; Gundersen, Y.; Hoke, A.V.; LeVangie, M.W.; Kumar, R.; Chakraborty, N.; et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am. J. Physiol. Liver Physiol. 2017, 312, G559–G571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, W.L.; Kalantar-Zadeh, K.; Vaziri, N.D. The Gut as a Source of Inflammation in Chronic Kidney Disease. Nephron 2015, 130, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.K.; Newman, V.L.; Romaine, P.L.; Hauer-Jensen, M.; Pollard, H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn. 2016, 16, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Bujold, K.; Hauer-Jensen, M.; Donini, O.; Rumage, A.; Hartman, D.; Hendrickson, H.P.; Stamatopoulos, J.; Naraghi, H.; Pouliot, M.; Ascah, A.; et al. Citrulline as a Biomarker for Gastrointestinal-Acute Radiation Syndrome: Species Differences and Experimental Condition Effects. Radiat. Res. 2016, 186, 71–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodionov, R.N.; Oppici, E.; Martens-Lobenhoffer, J.; Jarzebska, N.; Brilloff, S.; Burdin, D.; Demyanov, A.; Kolouschek, A.; Leiper, J.; Maas, R.; et al. A Novel Pathway for Metabolism of the Cardiovascular Risk Factor Homoarginine by alanine:glyoxylate aminotransferase 2. Sci. Rep. 2016, 6, 35277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, R.; Gangabhagirathi, R.; Venu, S.; Adhikari, S.; Mukherjee, T. Antioxidant activity and free radical scavenging reactions of gentisic acid: In-vitro and pulse radiolysis studies. Free. Radic. Res. 2011, 46, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ashidate, K.; Kawamura, M.; Mimura, D.; Tohda, H.; Miyazaki, S.; Teramoto, T.; Yamamoto, Y.; Hirata, Y. Gentisic acid, an aspirin metabolite, inhibits oxidation of low-density lipoprotein and the formation of cholesterol ester hydroperoxides in human plasma. Eur. J. Pharmacol. 2005, 513, 173–179. [Google Scholar] [CrossRef]
- Menni, C.; Zhu, J.; Le Roy, C.I.; Mompeo, O.; Young, K.; Rebholz, C.M.; Selvin, E.; North, K.E.; Mohney, R.P.; Bell, J.T.; et al. Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes 2020, 11, 1632–1642. [Google Scholar] [CrossRef]
- Lee, Y.; Khan, A.; Hong, S.; Jee, S.H.; Park, Y.H. A metabolomic study on high-risk stroke patients determines low levels of serum lysine metabolites: A retrospective cohort study. Mol. BioSyst. 2017, 13, 1109–1120. [Google Scholar] [CrossRef] [Green Version]
- Batra, V.; Kislay, B. Mitigation of gamma-radiation induced abasic sites in genomic DNA by dietary nicotinamide supplementation: Metabolic up-regulation of NAD+ biosynthesis. Mutat. Res. Mol. Mech. Mutagen. 2013, 749, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Rimmerman, N.; Bradshaw, H.B.; Hughes, H.V.; Chen, J.S.-C.; Hu, S.S.-J.; McHugh, D.; Vefring, E.; Jahnsen, J.A.; Thompson, E.L.; Masuda, K.; et al. N-Palmitoyl Glycine, a Novel Endogenous Lipid That Acts As a Modulator of Calcium Influx and Nitric Oxide Production in Sensory Neurons. Mol. Pharmacol. 2008, 74, 213–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, A.J.; Zhang, S.; Bhadelia, R.A.; Johnson, E.J.; Lichtenstein, A.H.; Rogers, G.T.; Rosenberg, I.H.; Smith, C.E.; Zeisel, S.H.; Scott, T.M. Choline and its metabolites are differently associated with cardiometabolic risk factors, history of cardiovascular disease, and MRI-documented cerebrovascular disease in older adults. Am. J. Clin. Nutr. 2017, 105, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Ueland, P.M. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2010, 34, 3–15. [Google Scholar] [CrossRef] [PubMed]






| Group | TP1 (Pre-Exposure) | TP2 | TP3 | TP4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day | D-5 a | D-1 b | D9 a | D10 b | D28 b | D29 a | D59 a | D60 b | ||||
| Sex | M | M | F | M | M | F | M | F | M | M | M | F |
| Sham | 4 | 1 | 2 | 4 | 2 | 2 | 1 | 2 | 4 | 4 | 2 | 2 |
| 8 Gy | 0 | 6 | 6 | 0 | 5 | 5 | 4 | 3 | 0 | 0 | 2 | 2 |
| 11 Gy | 12 | 6 | 6 | 12 | 6 | 5 | 4 | 3 | 11 | 10 | 1 | 1 |
| 12.5 Gy | 12 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 7 | 6 | 0 | 0 |
| 14 Gy | 0 | 6 | 6 | 0 | 3 | 4 | 2 | 1 | 0 | 0 | 2 | 0 |
| (A) | |||||||
| Radiation | TP1 vs. TP2 | TP1 vs. TP3 | TP1 vs. TP4 | ||||
| 0 Gy | 36 (0) | 54 (0) | 82 (6) | ||||
| 8 Gy | 12 (0) | 9 (0) | 25 (3) | ||||
| 11 Gy | 116 (112) | 86 (70) | 122 (116) | ||||
| 12.5 Gy | 173 (150) | 58 (11) | 94 (51) | ||||
| 14 Gy | 122 (29) | 34 (5) | 22 (0) | ||||
| (B) | |||||||
| Time | 0 Gy vs. 8 Gy | 0 Gy vs. 11 Gy | 0 Gy vs. 12.5 Gy | 0 Gy vs. 14 Gy | |||
| TP1 | 7 (0) | 6 (0) | 4 (0) | 2 (0) | |||
| TP2 | 37 (0) | 92 (64) | 154 (112) | 128 (34) | |||
| TP3 | 18 (0) | 25 (4) | 23 (2) | 51 (0) | |||
| TP4 | 43 (0) | 13 (0) | 26 (1) | 54 (4) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jun, S.-R.; Boerma, M.; Udaondo, Z.; Richardson, S.; Thrall, K.D.; Miousse, I.R.; Seng, J.; Pathak, R.; Hauer-Jensen, M. Plasma Metabolomics in a Nonhuman Primate Model of Abdominal Radiation Exposure. Metabolites 2021, 11, 540. https://doi.org/10.3390/metabo11080540
Jun S-R, Boerma M, Udaondo Z, Richardson S, Thrall KD, Miousse IR, Seng J, Pathak R, Hauer-Jensen M. Plasma Metabolomics in a Nonhuman Primate Model of Abdominal Radiation Exposure. Metabolites. 2021; 11(8):540. https://doi.org/10.3390/metabo11080540
Chicago/Turabian StyleJun, Se-Ran, Marjan Boerma, Zulema Udaondo, Sasha Richardson, Karla D. Thrall, Isabelle R. Miousse, John Seng, Rupak Pathak, and Martin Hauer-Jensen. 2021. "Plasma Metabolomics in a Nonhuman Primate Model of Abdominal Radiation Exposure" Metabolites 11, no. 8: 540. https://doi.org/10.3390/metabo11080540
APA StyleJun, S. -R., Boerma, M., Udaondo, Z., Richardson, S., Thrall, K. D., Miousse, I. R., Seng, J., Pathak, R., & Hauer-Jensen, M. (2021). Plasma Metabolomics in a Nonhuman Primate Model of Abdominal Radiation Exposure. Metabolites, 11(8), 540. https://doi.org/10.3390/metabo11080540