Postmortem Metabolomics: Strategies to Assess Time-Dependent Postmortem Changes of Diazepam, Nordiazepam, Morphine, Codeine, Mirtazapine and Citalopram
Abstract
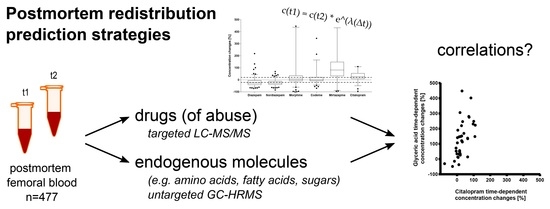
:1. Introduction
2. Results and Discussion
2.1. Storage and Shipping
2.2. Time-Dependent Concentration Changes of Drugs (of Abuse)
2.3. Strategies to Predict PMR/for a Posteriori Estimation of PMR Occurrence
2.3.1. Mixed Effect Models
2.3.2. Correlations with Endogenous Metabolites
3. Materials and Methods
3.1. Chemical and Reagents
3.2. Postmortem Sample Collection
3.3. Liquid Chromatography-Tandem Mass Spectrometric Analysis of Drugs (of Abuse)
3.4. Gas Chromatography-High Resolution Mass Spectrometric Analysis of Endogenous Compounds
3.5. Data Processing and Data Analysis
3.5.1. Drugs (of Abuse)
3.5.2. Endogenous Compounds
3.5.3. Mixed Effect Models
3.5.4. Correlation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skopp, G. Postmortem toxicology. Forensic Sci. Med. Pathol. 2010, 6, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Pelissier-Alicot, A.L.; Gaulier, J.M.; Champsaur, P.; Marquet, P. Mechanisms underlying postmortem redistribution of drugs: A review. J. Anal. Toxicol. 2003, 27, 533–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarema, M.C.; Becker, C.E. Key concepts in postmortem drug redistribution. Clin. Toxicol. 2005, 43, 235–241. [Google Scholar] [CrossRef]
- Lemaire, E.; Schmidt, C.; Denooz, R.; Charlier, C.; Boxho, P. Postmortem concentration and redistribution of diazepam, methadone, and morphine with subclavian and femoral vein dissection/clamping. J. Forensic Sci. 2016, 61, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, E.; Schmidt, C.; Denooz, R.; Charlier, C.; Boxho, P. Popliteal vein blood sampling and the postmortem redistribution of diazepam, methadone, and morphine. J. Forensic Sci. 2016, 61, 1017–1028. [Google Scholar] [CrossRef]
- Lemaire, E.; Schmidt, C.; Dubois, N.; Denooz, R.; Charlier, C.; Boxho, P. Site-, technique-, and time-related aspects of the postmortem redistribution of diazepam, methadone, morphine, and their metabolites: Interest of popliteal vein blood sampling. J. Forensic Sci 2017, 62, 1559–1574. [Google Scholar] [CrossRef]
- Giaginis, C.; Tsantili-Kakoulidou, A.; Theocharis, S. Quantitative structure-activity relationship (QSAR) methodology in forensic toxicology: Modeling postmortem redistribution of structurally diverse drugs using multivariate statistics. Forensic Sci. Int. 2009, 190, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Giaginis, C.; Tsantili-Kakoulidou, A.; Theocharis, S. Applying quantitative structure–activity relationship (QSAR) methodology for modeling postmortem redistribution of benzodiazepines and tricyclic antidepressants. J. Anal. Toxicol. 2014, 38, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Gerostamoulos, D.; Beyer, J.; Staikos, V.; Tayler, P.; Woodford, N.; Drummer, O.H. The effect of the postmortem interval on the redistribution of drugs: A comparison of mortuary admission and autopsy blood specimens. Forensic Sci. Med. Pathol. 2012, 8, 373–379. [Google Scholar] [CrossRef]
- Saar, E.; Beyer, J.; Gerostamoulos, D.; Drummer, O.H. The time-dependant post-mortem redistribution of antipsychotic drugs. Forensic Sci. Int. 2012, 222, 223–227. [Google Scholar] [CrossRef]
- Mantinieks, D.; Gerostamoulos, D.; Glowacki, L.; Di Rago, M.; Schumann, J.; Woodford, N.W.; Drummer, O.H. Postmortem drug redistribution: A compilation of postmortem/antemortem drug concentration ratios. J. Anal. Toxicol. 2021, 45, 368–377. [Google Scholar] [CrossRef]
- Staeheli, S.N.; Gascho, D.; Fornaro, J.; Laberke, P.; Ebert, L.C.; Martinez, R.M.; Thali, M.J.; Kraemer, T.; Steuer, A.E. Development of CT-guided biopsy sampling for time-dependent postmortem redistribution investigations in blood and alternative matrices--proof of concept and application on two cases. Anal. Bioanal. Chem. 2016, 408, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Staeheli, S.N.; Gascho, D.; Ebert, L.C.; Kraemer, T.; Steuer, A.E. Time-dependent postmortem redistribution of morphine and its metabolites in blood and alternative matrices-application of CT-guided biopsy sampling. Int. J. Legal Med. 2017, 131, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Brockbals, L.; Staeheli, S.N.; Gascho, D.; Ebert, L.C.; Kraemer, T.; Steuer, A.E. Time-dependent postmortem redistribution of opioids in blood and alternative matrices. J. Anal. Toxicol. 2018, 42, 365–374. [Google Scholar] [CrossRef]
- Brockbals, L.; Staeheli, S.N.; Gascho, D.; Ebert, L.C.; Kraemer, T.; Steuer, A.E. Time- and site-dependent postmortem redistribution of antidepressants and neuroleptics in blood and alternative matrices. J. Anal. Toxicol. 2021, 45, 356–367. [Google Scholar] [CrossRef]
- Langford, A.; Pounder, D. Possible markers for postmortem drug redistribution. J. Forensic Sci. 1997, 42, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Brockbals, L.; Staeheli, S.N.; Kraemer, T.; Steuer, A.E. Postmortem metabolomics: Correlating time-dependent concentration changes of xenobiotic and endogenous compounds. Drug Test. Anal. 2020, 12, 1171–1182. [Google Scholar] [CrossRef]
- Di Rago, M.; Pantatan, S.; Hargreaves, M.; Wong, K.; Mantinieks, D.; Kotsos, A.; Glowacki, L.; Drummer, O.H.; Gerostamoulos, D. High throughput detection of 327 drugs in blood by LC-MS-MS with automated data processing. J. Anal. Toxicol. 2021, 45, 154–183. [Google Scholar] [CrossRef]
- Staeheli, S.N.; Poetzsch, M.; Kraemer, T.; Steuer, A.E. Development and validation of a dynamic range-extended LC-MS/MS multi-analyte method for 11 different postmortem matrices for redistribution studies applying solvent calibration and additional 13C isotope monitoring. Anal. Bioanal. Chem. 2015, 407, 8681–8712. [Google Scholar] [CrossRef]
- Lee, J.E.; Kim, Y.Y. Impact of preanalytical variations in blood-derived biospecimens on omics studies: Toward precision biobanking? Omics 2017, 21, 499–508. [Google Scholar] [CrossRef]
- Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a standardized strategy of clinical metabolomics for the advancement of precision medicine. Metabolites 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerostamoulos, J.; Drummer, O.H. Postmortem redistribution of morphine and its metabolites. J. Forensic Sci. 2000, 45, 843–845. [Google Scholar] [CrossRef]
- Hargrove, V.M.; Molina, D.K. Peripheral postmortem redistribution of morphine. Am. J. Forensic Med. Pathol. 2014, 35, 106–108. [Google Scholar] [CrossRef]
- Langford, N.J.; Morley, S.R.; Ferner, R.E. The relationship between antemortem and postmortem morphine concentrations. Clin. Toxicol. 2019, 57, 1142–1145. [Google Scholar] [CrossRef]
- Moriya, F.; Hashimoto, Y. Distribution of free and conjugated morphine in body fluids and tissues in a fatal heroin overdose: Is conjugated morphine stable in postmortem specimens? J. Forensic Sci. 1997, 42, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Skopp, G.; Pötsch, L.; Klingmann, A.; Mattern, R. Stability of morphine, morphine-3-glucuronide, and morphine-6-glucuronide in fresh blood and plasma and postmortem blood samples. J. Anal. Toxicol. 2001, 25, 2–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maskell, P.D.; Albeishy, M.; De Paoli, G.; Wilson, N.E.; Seetohul, L.N. Postmortem redistribution of the heroin metabolites morphine and morphine-3-glucuronide in rabbits over 24 h. Int. J. Legal Med. 2016, 130, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Silk, M.J.; Harrison, X.A.; Hodgson, D.J. Perils and pitfalls of mixed-effects regression models in biology. PeerJ 2020, 8, e9522. [Google Scholar] [CrossRef]
- Stuart, B. Decomposition chemistry: Overview, analysis, and interpretation A2—Siegel, Jay A. In Encyclopedia of Forensic Sciences; Saukko, P.J., Houck, M.M., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 11–15. [Google Scholar]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, J.M.; Cascella, M. Physiology, enkephalin. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Brockbals, L.; Kraemer, T.; Steuer, A.E. Analytical considerations for postmortem metabolomics using GC-high-resolution MS. Anal. Bioanal. Chem. 2019, 412, 6241–6255. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Dowle, M.; Srinivasan, A. Data.Table: Extension of ‘Data.Frame’. R Package Version 1.14.0. Available online: https://CRAN.R-project.org/package=data.table (accessed on 15 February 2021).
- Auguie, B. gridExtra: Miscellaneous Functions for “Grid” Graphics. R Package Version 2.3. Available online: https://CRAN.R-project.org/package=gridExtra (accessed on 15 February 2021).
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Grolemund, G.; Wickham, H. Dates and times made easy with lubridate. J. Stat. Softw. 2011, 40, 1–25. [Google Scholar] [CrossRef]
- Komsta, L. outliers: Tests for Outliers. R Package Version 0.14. Available online: https://CRAN.R-project.org/package=outliers (accessed on 15 February 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; Francois, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]




| - | Pre-Admission interval (t0 − t1) [h] | Pre-Autopsy Interval (t0 − t2) [h] | ∆t (t1 − t2) [h] |
|---|---|---|---|
| Mean | 16 | 99 | 83 |
| Median | 9 | 86 | 70 |
| Min | 1.25 | 11 | 6.5 |
| Max | 292 | 478 | 434 |
| - | Diazepam | Nordiazepam | Morphine | Codeine | Mirtazapine | Citalopram |
|---|---|---|---|---|---|---|
| Lambda (λ) | −0.00296044 | −0.00303677 | 0.0014037 | 0.00020796 | 0.00333899 | 0.0037752 |
| RSD of λ [%] | 7.2 | 5.4 | 27 | 15 | ||
| Confidence interval (2.5–97.5%) | −0.00352471 to −0.00239861 | −0.00365846 to −0.00241503 | −0.00023068 to 0.00302557 | −0.00078685 to 0.00119634 | 0.00035995 to 0.00627887 | 0.00139422 to 0.00619429 |
| Median prediction accuracy [%] | 109 | 108 |
| Drug | Endogenous Compound/Feature | Spearman Correlation Coefficient | p-Value (FDR) | Workflow/ID Level |
|---|---|---|---|---|
| Diazepam n = 137 | Fumaric acid 2TMS | 0.38394 | <0.05 | untargeted/2 |
| Hexadecanoic acid (C16:0) TMS | 0.36859 | <0.05 | targeted/1 | |
| Oleic acid (C18:1) TMS | 0.35069 | <0.05 | untargeted/1 | |
| Glyceric acid 3TMS | −0.38241 | <0.05 | untargeted/1 | |
| Nordiazepam n = 126 | Fumaric acid 2TMS | 0.33849 | <0.05 | untargeted/2 |
| Ornithine 4TMS | 0.32831 | <0.05 | untargeted/1 | |
| Oleic acid (C18:1) TMS | 0.35069 | <0.05 | untargeted/1 | |
| Glyceric acid 3TMS | −0.46592 | <0.05 | targeted/1 | |
| 10.47_219.12279 | −0.32552 | <0.05 | untargeted/4 | |
| Morphine n = 122 | Methionine 2TMS | 0.57252 | <0.05 | targeted/1 |
| Phenylalanine 2TMS | 0.55928 | <0.05 | targeted/1 | |
| Valine 2TMS | 0.56490 | <0.05 | targeted/1 | |
| Isoleucine 2TMS | 0.52284 | <0.05 | targeted/1 | |
| Proline 2TMS | 0.51051 | <0.05 | targeted/1 | |
| Codeine n = 92 | Methionine 2TMS | 0.45389 | <0.05 | targeted/1 |
| Oleic acid (C18:1) TMS | 0.45302 | <0.05 | targeted/1 | |
| Phenylalanine 2TMS | 0.44358 | <0.05 | targeted/1 | |
| Valine 2TMS | 0.42247 | <0.05 | targeted/1 | |
| 11.679_245.10248 | −0.41774 | <0.05 | untargeted/4 | |
| Mirtazapine n = 55 | Methionine 2TMS | 0.58152 | <0.05 | targeted/1 |
| Uracil 2TMS | 0.57225 | <0.05 | targeted/1 | |
| Phenylalanine 2TMS | 0.56394 | <0.05 | targeted/1 | |
| Valine 2TMS | 0.51234 | <0.05 | targeted/1 | |
| Citalopram n = 50 | Glyceric acid 3TMS | 0.60497 | <0.05 | targeted/1 |
| Ribose 4TMS 1MOX | 0.55732 | <0.05 | targeted/1 | |
| Alanine 2TMS | 0.52508 | <0.05 | targeted/1 |
| Spectrum Properties Filter | |
| Lower retention time limit | 5.3 min |
| Upper retention time limit | 24.5 min |
| Peak Detection Settings | |
| Mass tolerance | 5 ppm |
| Spectral S/N threshold | 3 |
| Peak S/N threshold | 5 |
| Smoothing | 9 |
| TIC threshold | 500,000 |
| Ion overlap window | 98% |
| Group Compounds Settings | |
| Retention time tolerance | 10 s |
| Dot product threshold | 500 |
| Composition threshold | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brockbals, L.; Wartmann, Y.; Mantinieks, D.; Glowacki, L.L.; Gerostamoulos, D.; Kraemer, T.; Steuer, A.E. Postmortem Metabolomics: Strategies to Assess Time-Dependent Postmortem Changes of Diazepam, Nordiazepam, Morphine, Codeine, Mirtazapine and Citalopram. Metabolites 2021, 11, 643. https://doi.org/10.3390/metabo11090643
Brockbals L, Wartmann Y, Mantinieks D, Glowacki LL, Gerostamoulos D, Kraemer T, Steuer AE. Postmortem Metabolomics: Strategies to Assess Time-Dependent Postmortem Changes of Diazepam, Nordiazepam, Morphine, Codeine, Mirtazapine and Citalopram. Metabolites. 2021; 11(9):643. https://doi.org/10.3390/metabo11090643
Chicago/Turabian StyleBrockbals, Lana, Yannick Wartmann, Dylan Mantinieks, Linda L. Glowacki, Dimitri Gerostamoulos, Thomas Kraemer, and Andrea E. Steuer. 2021. "Postmortem Metabolomics: Strategies to Assess Time-Dependent Postmortem Changes of Diazepam, Nordiazepam, Morphine, Codeine, Mirtazapine and Citalopram" Metabolites 11, no. 9: 643. https://doi.org/10.3390/metabo11090643
APA StyleBrockbals, L., Wartmann, Y., Mantinieks, D., Glowacki, L. L., Gerostamoulos, D., Kraemer, T., & Steuer, A. E. (2021). Postmortem Metabolomics: Strategies to Assess Time-Dependent Postmortem Changes of Diazepam, Nordiazepam, Morphine, Codeine, Mirtazapine and Citalopram. Metabolites, 11(9), 643. https://doi.org/10.3390/metabo11090643