Metabolic Features of Saliva in Breast Cancer Patients
Abstract
:1. Introduction
2. Results
2.1. Metabolic Features of Saliva Composition in Breast Cancer
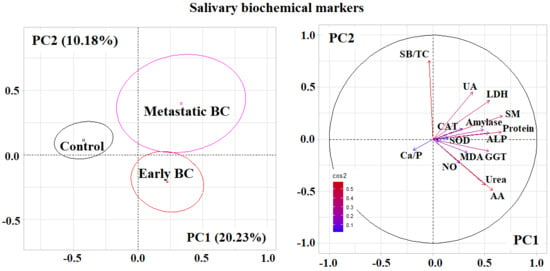
2.2. Metabolic Features of Saliva Composition Depending on the Stage of Breast Cancer
2.3. Metabolic Features of Saliva Composition Depending on the Histological Type of Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Study Design and Group Description
4.2. Collection, Processing, Storage and Analysis of Saliva Samples
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Kolyadina, I.V.; Poddubnaya, I.V.; Frank, G.A.; Komov, D.V.; Karseladze, A.I.; Ermilova, V.D.; Vishnevskaya, Y.V. Heterogenety of stage I breast cancer: Biological and prognostic value. Malig. Tumours 2015, 1, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Youlden, D.R.; Cramb, S.M.; Dunn, N.A.M.; Muller, J.M.; Pyke, C.M.; Baade, P.D. The descriptive epidemiology of female breast cancer: An international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012, 36, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Shah, R.; Rosso, K.; Nathanson, S.D. Pathogenesis, prevention, diagnosis and treatment of breast cancer. World J. Clin. Oncol. 2014, 5, 283–298. [Google Scholar] [CrossRef]
- Núñez, C. Blood-based protein biomarkers in breast cancer. Clin. Chim. Acta 2019, 490, 113–127. [Google Scholar] [CrossRef]
- Pak, D.D.; Rasskazova, E.A.; Danilova, T.V. Subtypes of breast cancer. Tumors Female Reprod. Syst. 2012, 3–4, 13–18. [Google Scholar]
- Wu, S.G.; He, Z.Y.; Zhou, J.; Sun, J.Y.; Li, F.Y.; Lin, Q.; Guo, L.; Lin, H.X. Serum levels of CEA and CA15-3 in different molecular subtypes and prognostic value in Chinese breast cancer. Breast 2014, 23, 88–93. [Google Scholar] [CrossRef]
- Uygur, M.M.; Gümüs, M. The utility of serum tumor markers CEA and CA 15–3 for breast cancer prognosis and their association with clinicopathological parameters. Cancer Treat. Res. Commun. 2021, 28, 100402. [Google Scholar] [CrossRef]
- Hing, J.X.; Mok, C.W.; Tan, P.T.; Sudhakar, S.S.; Seah, C.M.; Lee, W.P.; Tan, S.M. Clinical utility of tumour marker velocity of cancer antigen 15e3 (CA15e3) and carcinoembryonic antigen (CEA) in breast cancer surveillance. Breast 2020, 52, 95–101. [Google Scholar] [CrossRef]
- Marić, P.; Ozretić, P.; Levanat, S.; Oresković, S.; Antunac, K.; Beketić-Oresković, L. Tumor markers in breast cancer–evaluation of their clinical usefulness. Coll. Antropol. 2011, 35, 241–247. [Google Scholar]
- Hirata, B.K.B.; Oda, J.M.M.; Guembarovski, R.L.; Ariza, C.B.; de Oliveira, C.E.C.; Watanabe, M.A.E. Molecular markers for breast cancer: Prediction on tumor behavior. Dis. Markers 2014, 2014, 513158. [Google Scholar]
- Van Poznak, C.; Somerfield, M.R.; Bast, R.C.; Cristofanilli, M.; Goetz, M.P.; Gonzalez-Angulo, A.M.; Hicks, D.G.; Hill, E.G.; Liu, M.C.; Lucas, W.; et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American society of clinical oncology clinical practice guideline. J. Clin. Oncol. 2015, 33, 2695–2704. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.W.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. BBA Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef]
- Koopaie, M.; Abedinejad, F.; Manifar, S.; Mousavi, R.; Kolahdooz, S.; Shamshiri, A. Salivary miRNA-21 expression as a potential non-invasive diagnostic biomarker in breast cancer. Gene Rep. 2021, 25, 101317. [Google Scholar] [CrossRef]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; Canto, G.D.L.; Acevedo, A.C.; Guerra, E.N. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol. Hematol. 2017, 110, 62–73. [Google Scholar] [CrossRef]
- Navarro, M.A.; Mesia, R.; Diez-Gibert, O.; Rueda, A.; Ojeda, B.; Alonso, M.C. Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Cancer Res. Treat. 1997, 42, 83–86. [Google Scholar] [CrossRef]
- Pereira, D.d.A.; Areias, V.R.; Franco, M.F.; Benitez, M.C.; do Nascimento, C.M.; de Azevedo, C.M.; Alves, G. Measurement of HER2 in saliva of women in risk of breast cancer. Pathol. Oncol. Res. 2013, 19, 509–513. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L.; Tucci, M.; Thigpen, J.T. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Investig. 2000, 18, 101–109. [Google Scholar] [CrossRef]
- Brooks, M.N.; Wang, J.; Li, Y.; Zhang, R.; Elashoff, D.; Wong, D.T. Salivary protein factors are elevated in breast cancer patients. Mol. Med. Rep. 2008, 1, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Cavaco, C.; Pereira, J.A.M.; Taunk, K.; Taware, R.; Rapole, S.; Nagarajaram, H.; Câmara, J.S. Screening of salivary volatiles for putative breast cancer discrimination: An exploratory study involving geographically distant populations. Anal. Bioanal. Chem. 2018, 410, 4459–4468. [Google Scholar] [CrossRef]
- Al-Muhtaseb, S.I. Serum and saliva protein levels in females with breast cancer. Oncol. Lett. 2014, 8, 2752–2756. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, H.; Qiao, Y.; Yang, J.; Shu, J.; Zhang, J.; Zhang, Z.; He, J.; Li, Z. Salivary glycopatterns as potential biomarkers for screening of early-stage breast cancer. EBioMedicine 2018, 28, 70–79. [Google Scholar] [CrossRef] [Green Version]
- Murata, T.; Yanagisawa, T.; Kurihara, T.; Kaneko, M.; Ota, S.; Enomoto, A.; Tomita, M.; Sugimoto, M.; Sunamura, M.; Hayashida, T.; et al. Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res. Treat. 2019, 177, 591–601. [Google Scholar] [CrossRef]
- Pereira, J.A.M.; Taware, R.; Porto-Figueira, P.; Rapole, S.; Câmara, J.S. The salivary volatome in breast cancer. Precis. Med. Investig. Pract. Provid. 2020, 301–307. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Takayama, T.; Tsutsui, H.; Shimizu, I.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Inoue, K.; Todoroki, K.; Min, J.Z.; Mizuno, H.; et al. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin. Chim. Acta 2016, 452, 18–26. [Google Scholar] [CrossRef]
- Tsutsui, H.; Mochizuki, T.; Inoue, K.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Todoroki, K.; Min, J.Z.; Toyo’oka, T. High-throughput LC-MS/MS based simultaneous determination of polyamines including N-acetylated forms in human saliva and the diagnostic approach to breast cancer patients. Anal. Chem. 2013, 85, 11835–11842. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, Z.; Huang, Y.; Duan, Y.; Wang, X. Investigation of salivary free amino acid profile for early diagnosis of breast cancer with ultra-performance liquid chromatography-mass spectrometry. Clin. Chim. Acta 2015, 447, 23–31. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Kosenok, V.K. The level of cytokines in saliva in breast cancer. Vopr. Onkol. 2019, 65, 825–831. [Google Scholar]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Indicators of L-arginine metabolism in saliva: A focus on breast cancer. J. Oral Biosci. 2021, 63, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V.; Sarf, E.A. Determination of the content of organic acids in the saliva of patients with breast cancer by capillary electrophoresis. Klin. Lab. Diagn. 2018, 63, 419–422. [Google Scholar] [PubMed]
- Bel’skaya, L.V.; Sarf, E.A.; Shalygin, S.P.; Postnova, T.V.; Kosenok, V.K. Potential Diagnostic Significance of Salivary Copper Determination in Breast Cancer Patients: A Pilot Study. Biol. Trace Elem. Res. 2022, 200, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Delmonico, L.; Bravo, M.; Silvestre, R.T.; Ornellas, M.H.; De Azevedo, C.M.; Alves, G. Proteomic profile of saliva and plasma from women with impalpable breast lesions. Oncol. Lett. 2016, 12, 2145–2152. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J.; Shering, S.; Sherry, F.; McDermott, E.; O’Higgins, N. CA 15-3: A prognostic marker in breast cancer. Int J. Biol. Markers 2000, 15, 330–333. [Google Scholar] [CrossRef]
- Agha-Hosseini, F.; Mirzaii-Dizgah, I.; Rahimi, A.; Seilanian-Toosi, M. Correlation of serum and salivary CA125 levels in patients with breast cancer. J. Contemp Dent. Pract. 2009, 10, E001–E008. [Google Scholar]
- Farahani, H.; Amri, J.; Alaee, M.; Mohaghegh, F.; Rafiee, M. Serum and Saliva Levels of Cancer Antigen 15-3, Carcinoembryonic Antigen, Estradiol, Vaspin, and Obestatin as Biomarkers for the Diagnosis of Breast Cancer in Postmenopausal Women. Lab. Med. 2020, 51, 620–627. [Google Scholar] [CrossRef]
- Degnim, A.C.; Visscher, D.W.; Berman, H.K.; Frost, M.H.; Sellers, T.A.; Vierkant, R.A.; Maloney, S.D.; Pankratz, V.S.; de Groen, P.C.; Lingle, W.L.; et al. Stratification of breast cancer risk in women with atypia: A Mayo cohort study. J. Clin. Oncol. 2007, 25, 2671–2677. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K.; Gundyrev, I.A. Biochemical Markers of Saliva in Lung Cancer: Diagnostic and Prognostic Perspectives. Diagnostics 2020, 10, 186. [Google Scholar] [CrossRef] [Green Version]
- Emekli-Alturfan, E.; Demir, G.; Kasikci, E.; Tunali-Akbay, T.; Pisiriciler, R.; Caliskan, E.; Yarat, A. Altered biochemical parameters in the saliva of patients with breast cancer. Tohoku J. Exp. Med. 2008, 214, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Soukup, M.; Biesiada, I.; Henderson, A.; Idowu, B.; Rodeback, D.; Ridpath, L.; Bridges, E.G.; Nazar, A.M.; Bridges, K.G. Salivary uric acid as a noninvasive biomarker of metabolic syndrome. Diabetol. Metab. Syndr. 2012, 4, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gornitsky, M.; Velly, A.M.; Mohit, S.; Almajed, M.; Su, H.; Panasci, L.; Schipper, H.M. Altered levels of salivary 8-oxo-7-hydrodeoxyguanosine in breast cancer. JDR Clin. Transl. Res. 2016, 1, 171–177. [Google Scholar] [CrossRef]
- Hecht, F.; Pessoa, C.F.; Gentile, L.B.; Rosenthal, D.; Carvalho, D.P.; Fortunato, R.S. The role of oxidative stress on breast cancer development and therapy. Tumor Biol. 2016, 37, 4281–4291. [Google Scholar] [CrossRef]
- Gurer-Orhan, H.; Ince, E.; Konyar, D.; Saso, L.; Suzen, S. The role of oxidative stress modulators in breast cancer. Curr. Med. Chem. 2017, 25, 4084–4101. [Google Scholar] [CrossRef]
- Sawczuk, B.; Maciejczyk, M.; Sawczuk-Siemieniuk, M.; Posmyk, R.; Zalewska, A.; Car, H. Salivary Gland Function, Antioxidant Defence and Oxidative Damage in the Saliva of Patients with Breast Cancer: Does the BRCA1 Mutation Disturb the Salivary Redox Profile? Cancers 2019, 11, 1501. [Google Scholar] [CrossRef] [Green Version]
- Rajneesh, C.P.; Manimaran, A.; Sasikala, K.R.; Adaikappan, P. Lipid peroxidation and antioxidant status in patients with breast cancer. Singap. Med. J. 2008, 49, 640–643. [Google Scholar]
- Pande, D.; Negi, R.; Karki, K.; Khanna, S.; Khanna, R.S.; Khanna, H.D. Oxidative damage markers as possible discriminatory biomarkers in breast carcinoma. Transl. Res. 2012, 160, 411–418. [Google Scholar] [CrossRef]
- Knás, M.; Maciejczyk, M.; Waszkiel, D.; Zalewska, A. Oxidative stress and salivary antioxidants. Dent. Med. Probl. 2013, 50, 461–466. [Google Scholar]
- Liu, Y.; Zhou, Q.; Song, S.; Tang, S. Integrating metabolic reprogramming and metabolic imaging to predict breast cancer therapeutic responses. Trends Endocrinol. Metab. 2021, 32, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Abad, E.; Samino, S.; Yanes, O.; Potesil, D.; Zdrahal, Z.; Lyakhovich, A. Activation of glycogenolysis and glycolysis in breast cancer stem cell models. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165886. [Google Scholar] [CrossRef]
- Tsai, L.-C.; Hung, M.-W.; Chen, Y.-H.; Su, W.-C.; Chang, G.-G.; Chang, T.-C. Expression and regulation of alkaline phosphatases in human breast cancer MCF-7 cells. Eur. J. Biochem. 2000, 267, 1330–1339. [Google Scholar] [CrossRef]
- Moura, S.L.; Pallarès-Rusiñol, A.; Sappia, L.; Martí, M.; Pividori, M.I. The activity of alkaline phosphatase in breast cancer exosomes simplifies the biosensing design. Biosens. Bioelectron. 2022, 198, 113826. [Google Scholar] [CrossRef]
- Han, Y.; Park, I.S.; Kim, S.I.; Wang, W.; Yoo, J.; Jo, H.; Lee, J.; Seol, A.; Han, K.D.; Song, Y.S. Increasing serum gamma-glutamyltransferase level accompanies a rapid increase in the incidence of endometrial cancer in Korea: A nationwide cohort study. Gynecol. Oncol. 2021, 161, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Dawson, J.; Smith, G.D.; Boak, J.; Peters, T.J. γ-Glutamyltransferase in human and mouse breast tumours. Clin. Chim. Acta 1979, 96, 37–42. [Google Scholar] [CrossRef]
- Hanigan, M.H.; Frierson, H.F., Jr.; Swanson, P.E.; De Young, B.R. Altered expression of gamma-glutamyl transpeptidase in human tumors. Hum. Pathol. 1999, 30, 300–305. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Kosenok, V.K.; Massard, G. Activity of salivary metabolic enzymes in non-small cell lung cancer. Vopr. Onkol. 2017, 63, 926–932. [Google Scholar]
- Ramya, A.S.; Uppala, D.; Majumdar, S.; Surekha, C.; Deepak, K.G. Are salivary amylase and pH—Prognostic indicators of cancers? J. Oral Biol. Craniofacial Res. 2015, 5, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bel’skaya, L.V.; Kosenok, V.K.; Massard, G. Endogenous Intoxication and Saliva Lipid Peroxidation in Patients with Lung Cancer. Diagnostics 2016, 6, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]




| Indicator | Breast Cancer, n = 487 | Control Group, n = 298 | p-Value |
|---|---|---|---|
| Protein, g/L | 0.64 [0.37; 1.09] | 1.08 [0.65; 1.70] | 0.0000 |
| Urea, mmol/L | 9.63 [6.25; 13.38] | 6.66 [4.36; 9.13] | 0.0000 |
| Uric acid, μmol/L | 65.4 [24.1; 136.1] | 85.9 [34.4; 144.5] | 0.0119 |
| α-Aminoacids, mmol/L | 4.23 [3.89; 4.76] | 4.06 [3.83; 4.32] | 0.0000 |
| NO, μmol/L | 29.1 [17.4; 44.6] | 22.8 [13.2; 36.8] | 0.0001 |
| ALP, U/L | 72.8 [47.8; 106.5] | 60.8 [41.3; 84.7] | 0.0002 |
| LDH, U/L | 1451.0 [861.6; 2093.0] | 1101.5 [635.7; 1908.0] | 0.0002 |
| Catalase, nkat/mL | 3.78 [2.53; 5.99] | 4.58 [3.32; 5.79] | 0.0052 |
| MDA, μmol/L | 7.09 [5.81; 8.97] | 6.50 [5.73; 7.95] | 0.0006 |
| GGT, U/L | 23.2 [20.0; 26.5] | 20.4 [17.4; 24.4] | 0.0000 |
| SOD, c.u. | 73.7 [34.2; 142.1] | 57.9 [31.6; 113.2] | 0.0247 |
| α-Amylase, U/L | 306.5 [122.6; 605.3] | 185.2 [83.5; 384.4] | 0.0002 |
| Na/K-ratio, c.u. | 0.686 [0.481; 1.067] | 0.751 [0.505; 1.190] | 0.1270 |
| SOD/Catalase-ratio, c.u. | 19.8 [8.4; 39.7] | 14.5 [6.6; 37.7] | 0.0168 |
| SB/TC-ratio, c.u. | 0.627 [0.561; 0.704] | 0.609 [0.567; 0.667] | 0.0503 |
| Indicator | Group I, n = 226 | Group II, n = 131 | Group III, n = 75 | Group IV, n = 55 | Kruskal-Wallis Test (H, p) |
|---|---|---|---|---|---|
| Protein, g/L | 0.68 [0.39; 1.08] | 0.56 [0.37; 1.07] | 0.58 [0.29; 1.14] | 0.84 [0.46; 1.16] | 71.46; 0.0000 |
| Urea, mmol/L | 10.08 [6.66; 14.12] | 9.18 [5.90; 13.10] | 9.17 [6.00; 12.08] | 8.37 [5.83; 12.30] | 71.86; 0.0000 |
| Uric acid, μmol/L | 59.6 [23.2; 128.4] | 58.3 [26.3; 126.1] | 75.6 [16.7; 137.6] | 103.8 [39.5; 141.3] | 10.87; 0.0280 |
| α-Aminoacids, mmol/L | 4.29 [3.90; 4.89] | 4.23 [3.91; 4.67] | 4.14 [3.86; 4.72] | 4.24 [3.84; 4.65] | 25.64; 0.0000 |
| NO, μmol/L | 29.1 [17.2; 47.5] | 30.4 [18.2; 42.5] | 22.8 [13.9; 46.0] | 32.3 [22.1; 44.6] | 18.41; 0.0010 |
| ALP, U/L | 76.1 [50.0; 108.7] | 68.4 [47.8; 104.3] | 71.7 [41.3; 102.1] | 82.6 [47.8; 123.9] | 16.35; 0.0026 |
| LDH, U/L | 1509.0 [829.5; 2088.0] | 1452.5 [899.1; 2070.0] | 1280.0 [636.8; 2034.0] | 1703.0 [972.0; 2279.0] | 16.79; 0.0021 |
| Catalase, nkat/mL | 3.86 [2.65; 6.25] | 3.70 [2.36; 5.22] | 3.84 [2.25; 6.11] | 3.52 [2.56; 5.51] | 11.01; 0.0264 |
| MDA, μmol/L | 6.92 [5.73; 8.89] | 7.09 [5.90; 9.15] | 7.18 [5.94; 9.23] | 7.26 [5.81; 8.89] | 13.56; 0.0089 |
| GGT, U/L | 23.1 [19.8; 26.1] | 24.0 [20.4; 27.4] | 22.7 [19.9; 26.6] | 22.2 [19.4; 26.1] | 37.86; 0.0000 |
| Seromucoids, c.u. | 0.099 [0.062; 0.163] | 0.104 [0.068; 0.165] | 0.080 [0.055; 0.135] | 0.098 [0.055; 0.138] | 8.257; 0.0826 |
| SOD, c.u. | 76.3 [34.2; 168.4] | 57.9 [28.9; 113.2] | 75.0 [36.8; 134.2] | 73.7 [39.5; 113.2] | 10.31; 0.0355 |
| α-Amylase, U/L | 319.0 [157.7; 596.8] | 262.2 [124.8; 664.9] | 341.5 [111.6; 940.6] | 347.8 [115.4; 482.9] | 14.84; 0.0050 |
| Ca/P-ratio, c.u. | 0.256 [0.185; 0.354] | 0.274 [0.196; 0.380] | 0.274 [0.221; 0.450] | 0.266 [0.185; 0.345] | 10.40; 0.0343 |
| SB/TC-ratio, c.u. | 0.628 [0.557; 0.698] | 0.627 [0.566; 0.696] | 0.601 [0.556; 0.683] | 0.647 [0.569; 0.765] | 9.531; 0.0491 |
| Groups | n | UA/Urea, c.u. | UA·CAT/Urea, c.u. |
|---|---|---|---|
| Group I | 226 | 6.47 [1.97; 18.0] | 30.4 [6.80; 70.1] |
| <0.0001 | <0.0001 | ||
| Group II | 131 | 7.28 [2.15; 17.5] | 28.3 [5.50; 65.8] |
| <0.0001 | <0.0001 | ||
| Group III | 75 | 10.95 [1.33; 20.2] | 32.5 [3.70; 91.6] |
| 0.0026 | 0.0015 | ||
| Group IV | 55 | 11.78 [4.24; 20.3] | 42.6 [9.87; 92.2] |
| - | 0.0416 | ||
| Control | 298 | 15.05 [4.74; 29.2] | 63.3 [17.2; 145.9] |
| Indicator | Lobular BC, n = 86 | p-Value (Lobular vs. Control) | Ductal BC, n = 227 | p-Value (Ductal vs. Control) | p-Value (Ductal vs. Lobular) | Kruskal-Wallis Test (H, p) |
|---|---|---|---|---|---|---|
| Sodium, mmol/L | 6.5 [4.1; 11.8] | −22.2% 0.0435 | 7.7 [4.7; 12.5] | −7.8% 0.1455 | 0.2577 | 5.183; 0.0749 |
| Protein, g/L | 0.63 [0.36; 1.02] | −20.3% 0.0303 | 0.69 [0.40; 1.15] | −12.7% 0.1909 | 0.3301 | 49.97; 0.0000 * |
| Urea, mmol/L | 9.86 [4.86; 14.33] | +43.7% 0.0001 | 9.43 [6.03; 12.83] | +37.5% 0.0000 | 0.4063 | 41.49; 0.0000 * |
| Uric acid, μmol/L | 56.73 [27.03; 126.92] | −37.2% 0.0219 | 70.27 [25.00; 140.48] | −22.3% 0.0427 | 0.4440 | 4.922; 0.0853 |
| α-Aminoacids, mmol/L | 4.16 [3.89; 4.79] | +3.2% 0.0063 | 4.29 [3.88; 4.89] | +6.5% 0.0000 | 0.1737 | 20.75; 0.0000 * |
| NO, μmol/L | 25.4 [15.6; 44.6] | +8.0% 0.3540 | 26.8 [18.2; 40.7] | +14.0% 0.0355 | 0.6847 | 8.614; 0.0135 * |
| ALP, U/L | 69.54 [39.11; 97.79] | +10.3% 0.9190 | 73.88 [47.81; 108.65] | +17.2% 0.0055 | 0.0741 | 12.47; 0.0020 * |
| MM 254, c.u. | 0.206 [0.136; 0.332] | −21.1% 0.0064 | 0.256 [0.168; 0.382] | −1.9% 0.8093 | 0.0163 | 8.728; 0.0127 * |
| MM 280, c.u. | 0.177 [0.118; 0.291] | −19.2% 0.0139 | 0.200 [0.144; 0.342] | −8.7% 0.8621 | 0.0267 | 7.210; 0.0272 * |
| LDH, U/L | 1374.0 [731.8; 2008.0] | +27.7% 0.1590 | 1532.0 [1022.0; 2217.0] | +42.4% 0.0000 | 0.0852 | 20.12; 0.0000 * |
| Catalase, nkat/mL | 3.26 [2.45; 5.49] | −26.6% 0.0013 | 3.88 [2.52; 6.25] | −12.6% 0.0211 | 0.2193 | 7.107; 0.0286 * |
| TC, c.u. | 0.930 [0.822; 1.103] | +5.0% 0.0043 | 0.904 [0.800; 1.031] | +2.0% 0.0851 | 0.1197 | 3.821; 0.1480 |
| SB, c.u. | 0.576 [0.490; 0.755] | +7.3% 0.0004 | 0.555 [0.494; 0.686] | +3.4% 0.0009 | 0.2518 | 6.924; 0.0314 * |
| MDA, μmol/L | 6.92 [5.47; 8.29] | +3.7% 0.8404 | 7.14 [5.90; 9.15] | +7.1% 0.0181 | 0.1598 | 10.13; 0.0063 * |
| GGT, U/L | 21.9 [18.3; 24.9] | +5.3% 0.1069 | 23.4 [19.8; 26.5] | +12.5% 0.0000 | 0.0424 | 27.94; 0.0000 * |
| SOD, c.u. | 84.2 [31.6; 152.6] | +45.4% 0.0214 | 68.4 [39.5; 144.7] | +18.2% 0.0073 | 0.6601 | 6.430; 0.0402 * |
| α-Amylase, U/L | 217.4 [113.4; 451.5] | +17.4% 0.3211 | 304.7 [116.0; 526.4] | +64.5% 0.0077 | 0.3808 | 7.360; 0.0252 * |
| AOA, mmol/L | 2.46 [1.29; 3.52] | +4.2% 0.4735 | 2.23 [1.19; 3.18] | −5.5% 0.0205 | 0.4471 | 3.603; 0.1651 |
| Peroxidase, c.u. | 0.320 [0.210; 0.750] | −12.3% 0.9743 | 0.555 [0.290; 0.865] | +52.1% 0.0119 | 0.1040 | 4.026; 0.1336 |
| SOD/Catalase-ratio, c.u. | 21.5 [9.9; 41.7] | +50.5% 0.0052 | 19.9 [8.2; 42.8] | +39.3% 0.0010 | 0.6554 | 6.739; 0.0344 * |
| SB/(DC + TC)-ratio, c.u. | 0.115 [0.107; 0.153] | +3.6% 0.0006 | 0.112 [0.103; 0.137] | +0.9% 0.0140 | 0.1293 | 4.345; 0.1139 |
| SB/TC-ratio, c.u. | 0.644 [0.579; 0.707] | +6.8% 0.0040 | 0.631 [0.569; 0.723] | +4.6% 0.0006 | 0.8889 | 10.32; 0.0057 * |
| Feature | Breast Cancer, n = 487 | Control Group, n = 298 | |
|---|---|---|---|
| Age, years | 54.5 [47.0; 56.0] | 49.3 [43.8; 56.1] | |
| Histological type | |||
| Ductal | 227 (46.6%) | - | |
| Lobular | 86 (17.7%) | - | |
| Mixed (Ductal + Lobular) | 12 (2.5%) | - | |
| Rare forms | 58 (11.9%) | - | |
| Unknown | 104 (21.3%) | - | |
| Clinical Stage | |||
| Stage I | 119 (24.4%) | - | |
| Stage IIa | 123 (25.3%) | - | |
| Stage IIb | 88 (18.1%) | - | |
| Stage IIIa | 55 (11.3%) | - | |
| Stage IIIb | 47 (9.6%) | - | |
| Stage IV | 55 (11.3%) | - | |
| Subtype | |||
| Luminal A-like | 64 (13.1%) | - | |
| Luminal B-like (HER2+) | 230 (47.4%) | - | |
| Luminal B-like (HER2-) | 63 (12.9%) | - | |
| HER2-positive | 38 (7.8%) | - | |
| Triple-negative | 28 (5.7%) | - | |
| Unknown | 64 (13.1%) | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bel’skaya, L.V.; Sarf, E.A.; Solomatin, D.V.; Kosenok, V.K. Metabolic Features of Saliva in Breast Cancer Patients. Metabolites 2022, 12, 166. https://doi.org/10.3390/metabo12020166
Bel’skaya LV, Sarf EA, Solomatin DV, Kosenok VK. Metabolic Features of Saliva in Breast Cancer Patients. Metabolites. 2022; 12(2):166. https://doi.org/10.3390/metabo12020166
Chicago/Turabian StyleBel’skaya, Lyudmila V., Elena A. Sarf, Denis V. Solomatin, and Victor K. Kosenok. 2022. "Metabolic Features of Saliva in Breast Cancer Patients" Metabolites 12, no. 2: 166. https://doi.org/10.3390/metabo12020166
APA StyleBel’skaya, L. V., Sarf, E. A., Solomatin, D. V., & Kosenok, V. K. (2022). Metabolic Features of Saliva in Breast Cancer Patients. Metabolites, 12(2), 166. https://doi.org/10.3390/metabo12020166