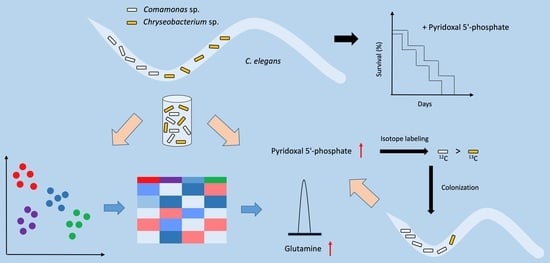
Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate
Abstract
:1. Introduction
2. Results
2.1. Presence and Abundance of PLP in the Native Bacterial Isolates
2.2. Presence and Abundance of PLP in the Worms Grown with the Native Bacterial Isolates
2.3. Contribution of PLP in the Worm from Each Individual Bacterium
2.4. Metabolome Profiling of Bacteria at the Global Level
2.5. Identification and Quantification of Glutamine
3. Discussion
4. Materials and Methods
4.1. Caenorhabditis Elegans and Bacterial Species
4.2. Worm Maintenance and Bacterial Culture
4.3. Detection and Measurement of PLP in Bacterial Isolates
4.4. Detection and Measurement of PLP in Worms Grown with the Native Bacterial Isolates or E. coli OP50
4.5. Isotope Labeling
4.6. Colonization Assay
4.7. Global Metabolomics of Bacteria
4.8. Detection and Measurement of Glutamine in Bacterial Isolates
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Dirksen, P.; Marsh, S.; Braker, I.; Heitland, N.; Wagner, S.; Nakad, R.; Mader, S.; Petersen, C.; Kowallik, V.; Rosenstiel, P.; et al. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol. 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirksen, P.; Assié, A.; Zimmermann, J.; Zhang, F.; Tietje, A.-M.; Marsh, S.A.; Félix, M.-A.; Shapira, M.; Kaleta, C.; Schulenburg, H.; et al. CeMbio—The Caenorhabditis elegans Microbiome Resource. G3 Genes Genomes Genet. 2020, 10, 3025–3039. [Google Scholar] [CrossRef] [PubMed]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, L.; Petersen, C.; Treitz, C.; Dierking, K.; Schulenburg, H.; Leippe, M.; Tholey, A. The Caenorhabditis elegans Proteome Response to Naturally Associated Microbiome Members of the Genus Ochrobactrum. Proteomics 2018, 18, e1700426. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Petersen, C.; Pees, B.; Zimmermann, J.; Waschina, S.; Dirksen, P.; Rosenstiel, P.; Tholey, A.; Leippe, M.; Dierking, K.; et al. The Inducible Response of the Nematode Caenorhabditis elegans to Members of Its Natural Microbiota Across Development and Adult Life. Front. Microbiol. 2019, 10, 1793. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Tateno, Y.; Gojobori, T. Evolution of Vitamin B6 (Pyridoxine) Metabolism by Gain and Loss of Genes. Mol. Biol. Evol. 2004, 22, 243–250. [Google Scholar] [CrossRef]
- Rosenberg, J.; Ischebeck, T.; Commichau, F.M. Vitamin B6 metabolism in microbes and approaches for fermentative production. Biotechnol. Adv. 2017, 35, 31–40. [Google Scholar] [CrossRef]
- Scott, T.; Quintaneiro, L.M.; Norvaisas, P.; Lui, P.; Wilson, M.; Leung, K.-Y.; Herrera-Dominguez, L.; Sudiwala, S.; Pessia, A.; Clayton, P.; et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 2017, 169, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Richts, B.; Rosenberg, J.; Commichau, F. A Survey of Pyridoxal 5′-Phosphate-Dependent Proteins in the Gram-Positive Model Bacterium Bacillus subtilis. Front. Mol. Biosci. 2019, 6, 32. [Google Scholar] [CrossRef]
- Barbier, T.; Collard, F.; Zúñiga-Ripa, A.; Moriyon, I.; Godard, T.; Becker, J.; Wittmann, C.; Van Schaftingen, E.; Letesson, J.-J. Erythritol feeds the pentose phosphate pathway via three new isomerases leading to D-erythrose-4-phosphate in Brucella. Proc. Natl. Acad. Sci. USA 2014, 111, 17815–17820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, J.; Obeng, N.; Yang, W.; Pees, B.; Petersen, C.; Waschina, S.; Kissoyan, K.A.; Aidley, J.; Hoeppner, M.P.; Bunk, B.; et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J. 2020, 14, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haçariz, O.; Viau, C.; Karimian, F.; Xia, J. The symbiotic relationship between Caenorhabditis elegans and members of its microbiome contributes to worm fitness and lifespan extension. BMC Genom. 2021, 22, 364. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zaiden, N.; Cao, B. The Core- and Pan-Genomic Analyses of the Genus Comamonas: From Environmental Adaptation to Potential Virulence. Front. Microbiol. 2018, 9, 3096. [Google Scholar] [CrossRef]
- Bachmann, T.; Maurer, A.; Rychlik, M. Development of a LC-MS/MS method using stable isotope dilution for the quantification of individual B6 vitamers in fruits, vegetables, and cereals. Anal. Bioanal. Chem. 2020, 412, 7237–7252. [Google Scholar] [CrossRef]
- Sajed, T.; Marcu, A.; Ramirez, M.; Pon, A.; Guo, A.C.; Knox, C.; Wilson, M.; Grant, J.R.; Djoumbou, Y.; Wishart, D.S. ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res. 2015, 44, D495–D501. [Google Scholar] [CrossRef] [Green Version]
- Metzler, B.; Gfeller, P.; Guinet, E. Restricting Glutamine or Glutamine-Dependent Purine and Pyrimidine Syntheses Promotes Human T Cells with High FOXP3 Expression and Regulatory Properties. J. Immunol. 2016, 196, 3618–3630. [Google Scholar] [CrossRef] [Green Version]
- De Vitto, H.; Arachchige, D.B.; Richardson, B.C.; French, J.B. The Intersection of Purine and Mitochondrial Metabolism in Cancer. Cells 2021, 10, 2603. [Google Scholar] [CrossRef]
- Elango, R. Methionine Nutrition and Metabolism: Insights from Animal Studies to Inform Human Nutrition. J. Nutr. 2020, 150, 2518S–2523S. [Google Scholar] [CrossRef]
- Leonardi, R.; Jackowski, S. Biosynthesis of Pantothenic Acid and Coenzyme A. EcoSal Plus 2007, 2, 687–694. [Google Scholar] [CrossRef] [Green Version]
- Genschel, U. Coenzyme A Biosynthesis: Reconstruction of the Pathway in Archaea and an Evolutionary Scenario Based on Comparative Genomics. Mol. Biol. Evol. 2004, 21, 1242–1251. [Google Scholar] [CrossRef] [Green Version]
- Neve, I.A.A.; Sowa, J.N.; Lin, C.-C.J.; Sivaramakrishnan, P.; Herman, C.; Ye, Y.; Han, L.; Wang, M.C. Escherichia coli Metabolite Profiling Leads to the Development of an RNA Interference Strain for Caenorhabditis elegans. G3 Genes|Genomes|Genet. 2020, 10, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Sivaramakrishnan, P.; Lin, C.-C.J.; Neve, I.A.; He, J.; Tay, L.W.R.; Sowa, J.N.; Sizovs, A.; Du, G.; Wang, J.; et al. Microbial Genetic Composition Tunes Host Longevity. Cell 2017, 169, 1249–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.Q.; Chong, J.; Zhou, G.Y.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.Z.; Xia, J.G. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purwaha, P.; Silva, L.P.; Hawke, D.H.; Weinstein, J.N.; Lorenzi, P.L. An Artifact in LC-MS/MS Measurement of Glutamine and Glutamic Acid: In-Source Cyclization to Pyroglutamic Acid. Anal. Chem. 2014, 86, 5633–5637. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 18 August 2020).






| KEGG Pathways | Hits (Total) | Hits (Sig) | p-Value (Gamma-Adjusted) |
|---|---|---|---|
| Purine metabolism | 24 | 8 | 0.02868 |
| Aminoacyl-tRNA biosynthesis | 17 | 7 | 0.01341 |
| Cysteine and methionine metabolism | 13 | 5 | 0.01839 |
| Pyrimidine metabolism | 12 | 6 | 0.00892 |
| Pantothenate and CoA biosynthesis | 11 | 6 | 0.00772 |
| Glutathione metabolism | 7 | 4 | 0.00872 |
| Lysine biosynthesis | 6 | 4 | 0.00726 |
| Glycine, serine and threonine metabolism | 6 | 3 | 0.01255 |
| Thiamine metabolism | 6 | 3 | 0.01255 |
| Histidine metabolism | 6 | 2 | 0.03089 |
| Sulfur metabolism | 6 | 2 | 0.03089 |
| Cyanoamino acid metabolism | 5 | 3 | 0.00949 |
| Tyrosine metabolism | 5 | 2 | 0.02224 |
| Riboflavin metabolism | 5 | 2 | 0.02224 |
| Purine metabolism (N) | 15 | 8 | 0.01759 |
| Galactose metabolism (N) | 6 | 3 | 0.0283 |
| Arginine and proline metabolism (N) | 6 | 3 | 0.0283 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haçariz, O.; Viau, C.; Gu, X.; Xia, J. Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate. Metabolites 2022, 12, 172. https://doi.org/10.3390/metabo12020172
Haçariz O, Viau C, Gu X, Xia J. Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate. Metabolites. 2022; 12(2):172. https://doi.org/10.3390/metabo12020172
Chicago/Turabian StyleHaçariz, Orçun, Charles Viau, Xue Gu, and Jianguo Xia. 2022. "Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate" Metabolites 12, no. 2: 172. https://doi.org/10.3390/metabo12020172
APA StyleHaçariz, O., Viau, C., Gu, X., & Xia, J. (2022). Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate. Metabolites, 12(2), 172. https://doi.org/10.3390/metabo12020172