Assessment of Tissue Specific Distribution and Seasonal Variation of Alkaloids in Alstonia scholaris
Abstract
:1. Introduction
2. Results
2.1. Identification of Alkaloids in AS
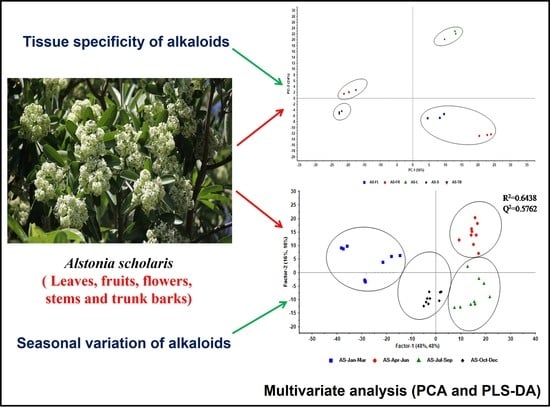
2.2. Tissue-Specific Profiling of Alkaloids in AS
2.3. Seasonal Variation of Alkaloids in AS
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Experimental Design
4.3. Metabolite Extraction
4.4. NMR Analysis
4.5. Quantitative Analysis
4.6. Multivariate Analysis (Data Reduction and Pattern Recognition)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baliga, M.S. Review of the Phytochemical, Pharmacological and Toxicological Properties of Alstonia Scholaris Linn. R. Br (Saptaparna). Chin. J. Integr. Med. 2012. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Adotey, J.P.K.; Adukpo, G.E.; Opoku Boahen, Y.; Armah, F.A. A Review of the Ethnobotany and Pharmacological Importance of Alstonia Boonei De Wild (Apocynaceae). ISRN Pharmacol. 2012, 2012, 587160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atta-Ur-Rahman, A.; Asif, M.; Ghazala, M.; Fatima, J.; Alvi, K.A. Scholaricine, an Alkaloid from Alstonia Scholaris. Phytochemistry 1985, 24, 2771–2773. [Google Scholar] [CrossRef]
- Guerriero, G.; Berni, R.; Muñoz-Sanchez, J.; Apone, F.; Abdel-Salam, E.; Qahtan, A.; Alatar, A.; Cantini, C.; Cai, G.; Hausman, J.-F.; et al. Production of Plant Secondary Metabolites: Examples, Tips and Suggestions for Biotechnologists. Genes 2018, 9, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, K.-H.; Araníbar, N.; Singh, B.; Stockton, G.W. Metabonomics Classifies Pathways Affected by Bioactive Compounds. Artificial Neural Network Classification of NMR Spectra of Plant Extracts. Phytochemistry 2003, 62, 971–985. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Nicholson, J.K.; Hylands, P.J.; Sampson, J.; Whitcombe, I.; Stewart, C.G.; Caiger, S.; Oru, I.; Holmes, E. Metabolomic Strategy for the Classification and Quality Control of Phytomedicine: A Case Study of Chamomile Flower (Matricaria Recutita L.). Planta Med. 2004, 70, 250–255. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, H.K.; Linthorst, H.J.M.; Hollander, J.G.; Lefeber, A.W.M.; Erkelens, C.; Nuzillard, J.-M.; Verpoorte, R. NMR Metabolomics to Revisit the Tobacco Mosaic Virus Infection in Nicotiana t Abacum Leaves. J. Nat. Prod. 2006, 69, 742–748. [Google Scholar] [CrossRef]
- Le Gall, G.; Colquhoun, I.J.; Defernez, M. Metabolite Profiling Using 1H NMR Spectroscopy for Quality Assessment of Green Tea, Camellia Sinensis (L.). J. Agric. Food Chem. 2004, 52, 692–700. [Google Scholar] [CrossRef]
- Pereira, G.E.; Gaudillere, J.-P.; van Leeuwen, C.; Hilbert, G.; Maucourt, M.; Deborde, C.; Moing, A.; Rolin, D. 1H NMR Metabolite Fingerprints of Grape Berry: Comparison of Vintage and Soil Effects in Bordeaux Grapevine Growing Areas. Anal. Chim. Acta 2006, 563, 346–352. [Google Scholar] [CrossRef]
- Schauer, N.; Semel, Y.; Roessner, U.; Gur, A.; Balbo, I.; Carrari, F.; Pleban, T.; Perez-Melis, A.; Bruedigam, C.; Kopka, J.; et al. Comprehensive Metabolic Profiling and Phenotyping of Interspecific Introgression Lines for Tomato Improvement. Nat. Biotechnol. 2006, 24, 447–454. [Google Scholar] [CrossRef]
- Graham, S.F.; Amigues, E.; Migaud, M.; Browne, R.A. Application of NMR Based Metabolomics for Mapping Metabolite Variation in European Wheat. Metabolomics 2009, 5, 302–306. [Google Scholar] [CrossRef]
- Brescia, M.A.; Caldarola, V.; De Giglio, A.; Benedetti, D.; Fanizzi, F.P.; Sacco, A. Characterization of the Geographical Origin of Italian Red Wines Based on Traditional and Nuclear Magnetic Resonance Spectrometric Determinations. Anal. Chim. Acta 2002, 458, 177–186. [Google Scholar] [CrossRef]
- Charlton, A.J.; Farrington, W.H.H.; Brereton, P. Application of 1H NMR and Multivariate Statistics for Screening Complex Mixtures: Quality Control and Authenticity of Instant Coffee. J. Agric. Food Chem. 2002, 50, 3098–3103. [Google Scholar] [CrossRef] [PubMed]
- Vogels, J.T.W.E.; Terwel, L.; Tas, A.C.; van den Berg, F.; Dukel, F.; van der Greef, J. Detection of Adulteration in Orange Juices by a New Screening Method Using Proton NMR Spectroscopy in Combination with Pattern Recognition Techniques. J. Agric. Food Chem. 1996, 44, 175–180. [Google Scholar] [CrossRef]
- Duarte, I.; Barros, A.; Belton, P.S.; Righelato, R.; Spraul, M.; Humpfer, E.; Gil, A.M. High-Resolution Nuclear Magnetic Resonance Spectroscopy and Multivariate Analysis for the Characterization of Beer. J. Agric. Food Chem. 2002, 50, 2475–2481. [Google Scholar] [CrossRef]
- Hussain, G.; Rasul, A.; Anwar, H.; Aziz, N.; Razzaq, A.; Wei, W.; Ali, M.; Li, J.; Li, X. Role of Plant Derived Alkaloids and Their Mechanism in Neurodegenerative Disorders. Int. J. Biol. Sci. 2018, 14, 341–357. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.-H.; Tan, Q.-G.; Liu, Y.-P.; Feng, T.; Du, Z.-Z.; Li, W.-Q.; Luo, X.-D. A Cage-Monoterpene Indole Alkaloid from Alstonia Scholaris. Org. Lett. 2008, 10, 577–580. [Google Scholar] [CrossRef]
- Pang, S.-Q.; Wang, G.-Q.; Lin, J.; Diao, Y.; Xu, R. Cytotoxic Activity of the Alkaloids from Broussonetia Papyrifera Fruits. Pharm. Biol. 2014, 52, 1315–1319. [Google Scholar] [CrossRef]
- Yang, X.-W.; Yang, C.-P.; Jiang, L.-P.; Qin, X.-J.; Liu, Y.-P.; Shen, Q.-S.; Chen, Y.-B.; Luo, X.-D. Indole Alkaloids with New Skeleton Activating Neural Stem Cells. Org. Lett. 2014, 16, 5808–5811. [Google Scholar] [CrossRef]
- Banerji, A.; Chakrabarty, M. Lochvinerine: A New Indole Alkaloid of Vinca Major. Phytochemistry 1974, 13, 2309–2312. [Google Scholar] [CrossRef]
- Caron, C.; Graftieaux, A.; Massiot, G.; Le Men-Olivier, L.; Delaude, C. Alkaloids from Alstonia Congensis. Phytochemistry 1989, 28, 1241–1244. [Google Scholar] [CrossRef]
- Yamauchi, T.; Abe, F.; Padolina, W.G.; Dayrit, F.M. Alkaloids from Leaves and Bark of Alstonia Scholaris in the Philippines. Phytochemistry 1990, 29, 3321–3325. [Google Scholar] [CrossRef]
- Bhattacharya, S.K.; Bose, R.; Dutta, S.C.; Ray, A.B.; Guha, S.R. Neuropharmacological Studies on Strictamine Isolated from Alstonia Scholaris. Indian J. Exp. Biol. 1979, 17, 598–600. [Google Scholar] [PubMed]
- Morita, Y.; Hesse, M.; Schmid, H.; Banerji, A.; Banerji, J.; Chatterjee, A.; Oberhänsli, W.E. Alstonia Scholaris: Struktur Des Indolalkaloides Narelin. HCA 1977, 60, 1419–1434. [Google Scholar] [CrossRef]
- Boonchuay, W.; Court, W. Alkaloids of Alstonia Scholaris from Thailand. Planta Med. 1976, 29, 380–390. [Google Scholar] [CrossRef]
- Rastogi, R.C.; Kapil, R.S.; Popli, S.P. Picralinal—A Key Alkaloid of Picralima Group from Alstonia Scholaris R. Br. Experientia 1970, 26, 1056. [Google Scholar] [CrossRef] [Green Version]
- Macabeo, A.P.G.; Krohn, K.; Gehle, D.; Read, R.W.; Brophy, J.J.; Cordell, G.A.; Franzblau, S.G.; Aguinaldo, A.M. Indole Alkaloids from the Leaves of Philippine Alstonia scholaris. Phytochemistry 2005, 66, 1158–1162. [Google Scholar] [CrossRef]
- Toh-Seok, K.; Kok-Tih, N.; Kooi-Mow, S.; Yoganathan, K. Alkaloids from Alstonia scholaris. Phytochemistry 1997, 45, 1303–1305. [Google Scholar] [CrossRef]
- Salim, A.A.; Garson, M.J.; Craik, D.J. New Indole Alkaloids from the Bark of Alstonia scholaris. J. Nat. Prod. 2004, 67, 1591–1594. [Google Scholar] [CrossRef]
- Thomford, N.; Senthebane, D.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef] [Green Version]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, E37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Zhang, Z.; Li, S.; Ye, X.; Li, X.; He, K. Synergy Effects of Herb Extracts: Pharmacokinetics and Pharmacodynamic Basis. Fitoterapia 2014, 92, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Grkovic, T.; Pouwer, R.H.; Vial, M.-L.; Gambini, L.; Noël, A.; Hooper, J.N.A.; Wood, S.A.; Mellick, G.D.; Quinn, R.J. NMR Fingerprints of the Drug-like Natural-Product Space Identify Iotrochotazine A: A Chemical Probe to Study Parkinson’s Disease. Angew. Chem. Int. Ed. 2014, 53, 6070–6074. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.-S.; Short, L.C.; Syage, J.A.; Potvin, M.; Curtis, J.M. Liquid Chromatography–Atmospheric Pressure Photoionization-Mass Spectrometry Analysis of Triacylglycerol Lipids—Effects of Mobile Phases on Sensitivity. J. Chromatogr. A 2007, 1173, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Dighe, S.U.; Mahar, R.; Shukla, S.K.; Kant, R.; Srivastava, K.; Batra, S. Synthesis of S-(−)-5,6-Dihydrocanthin-4-Ones via a Triple Cooperative Catalysis-Mediated Domino Reaction. J. Org. Chem. 2016, 81, 4751–4761. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, W.F. Natural Product Structure Elucidation by NMR Spectroscopy. In Pharmacognosy; Elsevier: Amsterdam, The Netherlands, 2017; pp. 567–596. ISBN 978-0-12-802104-0. [Google Scholar]
- Pandey, R.; Mahar, R.; Hasanain, M.; Shukla, S.K.; Sarkar, J.; Rameshkumar, K.B.; Kumar, B. Rapid Screening and Quantitative Determination of Bioactive Compounds from Fruit Extracts of Myristica Species and Their in Vitro Antiproliferative Activity. Food Chem. 2016, 211, 483–493. [Google Scholar] [CrossRef]
- Yang, L.; Stöckigt, J. Trends for Diverse Production Strategies of Plant Medicinal Alkaloids. Nat. Prod. Rep. 2010, 27, 1469. [Google Scholar] [CrossRef]
- Ward, J.L.; Baker, J.M.; Beale, M.H. Recent Applications of NMR Spectroscopy in Plant Metabolomics: NMR Spectroscopy in Plant Metabolomics. FEBS J. 2007, 274, 1126–1131. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-Based Metabolomic Analysis of Plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Masetti, O.; Nisini, L.; Ciampa, A.; Dell’Abate, M.T. 1H NMR Spectroscopy Coupled with Multivariate Analysis Was Applied to Investigate Italian Cherry Tomatoes Metabolic Profile. J. Chemom. 2020, 34, e3191. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. CMB 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Abdel-Farid, I.B.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Metabolic Characterization of Brassica Rapa Leaves by NMR Spectroscopy. J. Agric. Food Chem. 2007, 55, 7936–7943. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Tohge, T. The Genetics of Plant Metabolism. Annu. Rev. Genet. 2017, 51, 287–310. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, R.B. Plants: Novel Developmental Processes. Science 1988, 240, 1460–1467. [Google Scholar] [CrossRef]
- Cook, D.; Manson, J.S.; Gardner, D.R.; Welch, K.D.; Irwin, R.E. Norditerpene Alkaloid Concentrations in Tissues and Floral Rewards of Larkspurs and Impacts on Pollinators. Biochem. Syst. Ecol. 2013, 48, 123–131. [Google Scholar] [CrossRef]
- Prinsloo, G.; Nogemane, N. The Effects of Season and Water Availability on Chemical Composition, Secondary Metabolites and Biological Activity in Plants. Phytochem. Rev. 2018, 17, 889–902. [Google Scholar] [CrossRef]
- Zhou, H.; He, H.-P.; Luo, X.-D.; Wang, Y.-H.; Yang, X.-W.; Di, Y.-T.; Hao, X.-J. Three New Indole Alkaloids from the Leaves of Alstonia Scholaris. HCA 2005, 88, 2508–2512. [Google Scholar] [CrossRef]
- Feng, T.; Li, Y.; Cai, X.-H.; Gong, X.; Liu, Y.-P.; Zhang, R.-T.; Zhang, X.-Y.; Tan, Q.-G.; Luo, X.-D. Monoterpenoid Indole Alkaloids from Alstonia Yunnanensis. J. Nat. Prod. 2009, 72, 1836–1841. [Google Scholar] [CrossRef]
- Bharti, S.K.; Sinha, N.; Joshi, B.S.; Mandal, S.K.; Roy, R.; Khetrapal, C.L. Improved Quantification from 1H-NMR Spectra Using Reduced Repetition Times. Metabolomics 2008, 4, 367–376. [Google Scholar] [CrossRef]
- Sethi, N.; Mahar, R.; Shukla, S.K.; Kumar, A.; Sinha, N. A Novel Approach for Testing the Teratogenic Potential of Chemicals on the Platform of Metabolomics: Studies Employing HR-MAS Nuclear Magnetic Resonance Spectroscopy. RSC Adv. 2015, 5, 26027–26039. [Google Scholar] [CrossRef]
- Paul, A.; Kumar, S.; Raj, A.; Sonkar, A.A.; Jain, S.; Singhai, A.; Roy, R. Alteration in Lipid Composition Differentiates Breast Cancer Tissues: A 1H HRMAS NMR Metabolomic Study. Metabolomics 2018, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, F.; Ferro, M.; Mavroudakis, E.; Pellitteri, R.; Bossolasco, P.; Zaccheo, D.; Morbidelli, M.; Silani, V.; Mele, A.; Moscatelli, D.; et al. NMR Metabolomics for Stem Cell Type Discrimination. Sci. Rep. 2017, 7, 15808. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| S. No. | Compounds | 1H Chemical Shift (ppm) | 13C Chemical Shift (ppm) |
|---|---|---|---|
| 1 | Picrinine | 7.13 (d, J = 8.1 Hz), 7.07 (t, J = 8.0 Hz), 6.85 (d, J = 8.1 Hz), 6.78 (t, J = 8.0 Hz), 5.40 (q), 4.85 (m), 3.65 (s), 3.41 (m), 3.30 (m), 2.44 (d, J = 3.1 Hz), 2.24 (m), 2.13 (m), 1.50, 1.85 (m) | 172.4, 127, 125, 121.0, 120, 110, 87.3, 51.7, 51.4, 51.3, 40.6, 31.1, 26.1, 12.7 |
| 2 | Picralinal | 8.56 (s), 7.32 (d, J = 7.5 Hz), 7.11 (t, J = 7.4 Hz), 6.88 (t, J = 7.4 Hz), 6.70 (d, J = 7.5 Hz), 5.40 (q), 3.80 (m), 3.69 (s), 3.67 (brs), 3.54 (m), 3.47 (m), 3.11 (m), 2.24 (m), 1.98 (m), 1.50 (m) | 197.7, 168.0, 128.7, 126.l, 121.5, 121.0, 110.7, 65.1, 53.6, 52.5, 46.4, 43.1, 31.4, 22.6, 12.8 |
| 3 | Akuammidine | 7.53 (d, J = 7.6 Hz), 7.11 (d, J = 7.6 Hz), 7.04 (t, J = 7.5 Hz), 6.78 (t, J = 7.5 Hz), 5.38 (brq), 3.84 (m), 3.68 (m), 2.94 (s), 2.66 (m), 2.29 (m), 1.86 (m), 1.66 (d, J = 5.0 Hz) | 173.8, 121.5, 119.4, 118.1, 116.7, 110.9, 65.7, 51.3, 51.0, 39.5, 29.2, 13.0 |
| 4 | 19 S Scholaricine | 8.62 (brs), 7.48 (d, J = 7.6 Hz), 7.35 (d, J = 7.6 Hz), 7.04 (t, J = 7.6 Hz), 3.88 (s), 3.28 (m), 3.1 (m), 2.02 (m), 1.72 (m), 1.17 (m) | 168.8, 141.2, 122.5, 119.4, 118.1, 96.8, 68.5, 52.1, 31.0, 29.4, 12.5 |
| 5 | 19,20 E Vallesamine | 7.67 (d, J = 7.6 Hz), 7.30 (d, J = 7.6 Hz), 7.17 (t, J = 7.6 Hz), 7.07 (t, J = 7.6 Hz), 5.55 (q), 4.84 (m), 4.21 (d, J = 10.8 Hz), 3.75 (s), 3.63 (m), 2.33 (m), 1.90 (m), 1.74 (m) | 174.5, 123.0, 122.5, 119.4, 118.1, 110.0, 70.4, 58.5, 53.0, 36.2, 23.9, 14.1 |
| 6 | Nb-Demethylalstogustine N-Oxide | 7.67 (d, J = 7.5 Hz), 7.41 (t, J = 7.5 Hz), 6.97 (t, J = 7.5 Hz), 6.86 (d, J = 7.5 Hz), 4.18 (q), 3.80 (s), 3.44 (m), 3.20 (m), 1.38 (m), 1.29 (m) | 167.2, 129.1, 121.5, 120.1, 110.0, 103.7, 68.6, 51.3, 24.8, 24.0, 20.3 |
| 7 | Nb-Demethylalstogustine | 7.25 (d, J = 7.6 Hz), 7.18 (t, J = 7.6 Hz), 6.93 (t, J = 7.6 Hz), 6.86 (d, J = 7.6 Hz), 4.05, 3.84 (s), 3.09 (m), 2.35 (m), 1.85, 1.49 (m), 1.24 (m) | 167.5, 129.1, 122.5, 121.1, 110.2, 103.1, 68.0, 51.5, 44.4, 28.5, 26.1, 12.7 |
| 8 | Echitamine | 7.74 (d, J = 7.5 Hz), 7.72 (d, J = 7.5 Hz), 7.40 (t, J = 7.5 Hz), 6.25 (t, J = 7.5 Hz), 5.73 (q), 3.73 (s), 3.29 (s), 3.16 (m), 2.24 (m), 2.02 (m) | 171.0, 157.3, 129.8, 128.7, 126.7, 119.5, 110.6, 66.3, 64.5, 51.8, 29.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahar, R.; Manivel, N.; Kanojiya, S.; Mishra, D.K.; Shukla, S.K. Assessment of Tissue Specific Distribution and Seasonal Variation of Alkaloids in Alstonia scholaris. Metabolites 2022, 12, 607. https://doi.org/10.3390/metabo12070607
Mahar R, Manivel N, Kanojiya S, Mishra DK, Shukla SK. Assessment of Tissue Specific Distribution and Seasonal Variation of Alkaloids in Alstonia scholaris. Metabolites. 2022; 12(7):607. https://doi.org/10.3390/metabo12070607
Chicago/Turabian StyleMahar, Rohit, Nagarajan Manivel, Sanjeev Kanojiya, Dipak K. Mishra, and Sanjeev K. Shukla. 2022. "Assessment of Tissue Specific Distribution and Seasonal Variation of Alkaloids in Alstonia scholaris" Metabolites 12, no. 7: 607. https://doi.org/10.3390/metabo12070607
APA StyleMahar, R., Manivel, N., Kanojiya, S., Mishra, D. K., & Shukla, S. K. (2022). Assessment of Tissue Specific Distribution and Seasonal Variation of Alkaloids in Alstonia scholaris. Metabolites, 12(7), 607. https://doi.org/10.3390/metabo12070607