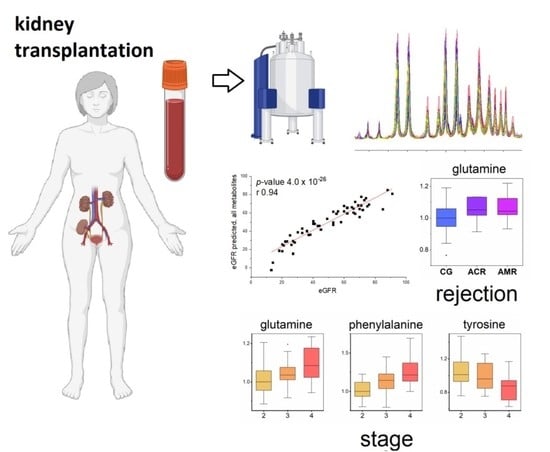
Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
- Age > 18 years,
- No active infection,
- Immunosuppression consisted of TAC, MPA, and Prednisone,
- Primary kidney transplantation from SCD.
- Age < 18 years,
- Lost to follow up,
- Active infection,
- Kidney transplantation from an extended criteria donor (ECD) or living kidney donor,
- Kidney transplantation in the last 6 months.
2.2. Samples Preparation
2.3. Data Acquisition
2.4. Data Processing
3. Results
4. Discussion
4.1. Alterations in Plasma Metabolites
4.2. Circulating Metabolites as Predictors of Allograft Rejection
4.3. Circulating Metabolites as Predictors of Allograft Function
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van de Poll, M.C.; Soeters, P.B.; Deutz, N.E.; Fearon, K.C.; Dejong, C.H. Renal metabolism of amino acids: Its role in interorgan amino acid exchange. Am. J. Clin. Nutr. 2004, 79, 185–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metabolic Adaptation of the Kidney to Hyperammonemia during Chronic Liver Insufficiency in the rat—Dejong—1993—Hepatology—Wiley Online Library. Available online: https://aasldpubs.onlinelibrary.wiley.com/doi/abs/10.1002/hep.1840180422 (accessed on 17 March 2022).
- Ammonia and Glutamine Metabolism During Liver Insufficiency: The Role of Kidney and Brain in Interorgan Nitrogen Exchange. Available online: https://www.tandfonline.com/doi/abs/10.3109/00365529609094733 (accessed on 17 March 2022).
- Wijermars, L.G.M.; Schaapherder, A.F.; de Vries, D.K.; Verschuren, L.; Wüst, R.C.I.; Kostidis, S.; Mayboroda, O.A.; Prins, F.; Ringers, J.; Bierau, J.; et al. Defective postreperfusion metabolic recovery directly associates with incident delayed graft function. Kidney Int. 2016, 90, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Stenlund, H.; Madsen, R.; Vivi, A.; Calderisi, M.; Lundstedt, T.; Tassini, M.; Carmellini, M.; Trygg, J. Monitoring kidney-transplant patients using metabolomics and dynamic modeling. Chemom. Intell. Lab. Syst. 2009, 98, 45–50. [Google Scholar] [CrossRef]
- Suhre, K.; Schwartz, J.E.; Sharma, V.K.; Chen, Q.; Lee, J.R.; Muthukumar, T.; Dadhania, D.M.; Ding, R.; Ikle, D.N.; Bridges, N.D.; et al. Urine Metabolite Profiles Predictive of Human Kidney Allograft Status. J. Am. Soc. Nephrol. 2016, 27, 626–636. [Google Scholar] [CrossRef] [Green Version]
- Calderisi, M.; Vivi, A.; Mlynarz, P.; Tassin, M.; Banasik, M.; Dawiskiba, T.; Carmellini, M. Using Metabolomics to Monitor Kidney Transplantation Patients by Means of Clustering to Spot Anomalous Patient Behavior. Transplant. Proc. 2013, 45, 1511–1515. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Gowda, Y.N.; Raftery, D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal. Chem. 2015, 87, 706–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Kruskal Wallis Test Calculator—With Post-Hoc Dunn’s Test Multiple Comparisons. Available online: https://www.statskingdom.com/kruskal-wallis-calculator.html (accessed on 16 March 2022).
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Møller, N.; Meek, S.; Bigelow, M.; Andrews, J.; Nair, K.S. The kidney is an important site for in vivo phenylalanine-to-tyrosine conversion in adult humans: A metabolic role of the kidney. Proc. Natl. Acad. Sci. USA 2000, 97, 1242–1246. [Google Scholar] [CrossRef] [Green Version]
- Kopple, J.D. Phenylalanine and Tyrosine Metabolism in Chronic Kidney Failure. J. Nutr. 2007, 137, 1586S–1590S. [Google Scholar] [CrossRef]
- Boirie, Y.; Albright, R.; Bigelow, M.; Nair, K.S. Impairment of phenylalanine conversion to tyrosine inend-stage renal disease causing tyrosine deficiency. Kidney Int. 2004, 66, 591–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newsholme, P.; Curi, R.; Pithon Curi, T.C.; Murphy, C.J.; Garcia, C.; Pires de Melo, M. Glutamine metabolism by lymphocytes, macrophages, and neutrophils: Its importance in health and disease. J. Nutr. Biochem. 1999, 10, 316–324. [Google Scholar] [CrossRef]
- Calder, P.C.; Yaqoob, P. Glutamine and the immune system. Amino Acids 1999, 17, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Cruzat, V.; Rogero, M.M.; Keane, K.N.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.; Curthoys, N.P. Glutamine metabolism: Role in acid-base balance. Biochem. Mol. Biol. Educ. 2004, 32, 291–304. [Google Scholar] [CrossRef]
- Weiner, I.D.; Verlander, J.W. Renal Ammonia Metabolism and Transport. Compr. Physiol. 2013, 3, 201. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Abramowitz, M.K. Metabolic acidosis and the progression of chronic kidney disease. BMC Nephrol. 2014, 15, 55. [Google Scholar] [CrossRef] [Green Version]
- Ritter, A.; Mohebbi, N. Causes and Consequences of Metabolic Acidosis in Patients after Kidney Transplantation. Kidney Blood Press. Res. 2020, 45, 792–801. [Google Scholar] [CrossRef]
- Adamczak, M.; Masajtis-Zagajewska, A.; Mazanowska, O.; Madziarska, K.; Stompór, T.; Więcek, A. Diagnosis and Treatment of Metabolic Acidosis in Patients with Chronic Kidney Disease—Position Statement of the Working Group of the Polish Society of Nephrology. Kidney Blood Press. Res. 2018, 43, 959–969. [Google Scholar] [CrossRef] [Green Version]
- May, R.C.; Kelly, R.A.; Mitch, W.E. Metabolic acidosis stimulates protein degradation in rat muscle by a glucocorticoid-dependent mechanism. J. Clin. Investig. 1986, 77, 614–621. [Google Scholar] [CrossRef] [Green Version]
- Tizianello, A.; De Ferrari, G.; Garibotto, G.; Robaudo, C. Amino acid metabolism and the liver in renal failure. Am. J. Clin. Nutr. 1980, 33, 1354–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppe, L.; Cassani de Oliveira, M.; Fouque, D. Ketoacid Analogues Supplementation in Chronic Kidney Disease and Future Perspectives. Nutrients 2019, 11, 2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.-B.; Cheng, B.-C.; Kao, T.-W. A comparison of progression of chronic renal failure: Low dose vs. standard dose ketoacIDS. Kidney Res. Clin. Pract. 2012, 31, A24. [Google Scholar] [CrossRef] [Green Version]
- Vera-Aviles, M.; Vantana, E.; Kardinasari, E.; Koh, N.L.; Latunde-Dada, G.O. Protective Role of Histidine Supplementation Against Oxidative Stress Damage in the Management of Anemia of Chronic Kidney Disease. Pharmaceuticals 2018, 11, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holeček, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Liu, T.; Yin, M. Beneficial effects of histidine and carnosine on ethanol-induced chronic liver injury. Food Chem. Toxicol. 2008, 46, 1503–1509. [Google Scholar] [CrossRef]
- Cueto-Manzano, A.M.; Morales-Buenrostro, L.E.; González-Espinoza, L.; González-Tableros, N.; Martín-del-Campo, F.; Correa-Rotter, R.; Valera, I.; Alberú, J. Markers of Inflammation before and after Renal Transplantation. Transplantation 2005, 80, 47–51. [Google Scholar] [CrossRef]
- Branco, A.C.C.C.; Yoshikawa, F.S.Y.; Pietrobon, A.J.; Sato, M.N. Role of Histamine in Modulating the Immune Response and Inflammation. Mediators Inflamm. 2018, 2018, e9524075. [Google Scholar] [CrossRef]
- Hamm, L.L. Renal handling of citrate. Kidney Int. 1990, 38, 728–735. [Google Scholar] [CrossRef] [Green Version]
- Hering-Smith, K.S.; Hamm, L.L. Acidosis and citrate: Provocative interactions. Ann. Transl. Med. 2018, 6, 29. [Google Scholar] [CrossRef]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calder, P.C. Branched-Chain Amino Acids and Immunity. J. Nutr. 2006, 136, 288S–293S. [Google Scholar] [CrossRef] [PubMed]





| Parameter | All | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 |
|---|---|---|---|---|---|---|
| Sample size | 55 | 1 | 21 | 21 | 10 | 2 |
| Age/years (SD) | 52.6 (13.8) | 41 | 48.4 (13.6) | 56.8 (13.7) | 54.4 (12.7) | 42, 48 (2 values) |
| Sex (F/M) | 23/32 | 0/1 | 10/11 | 8/13 | 5/5 | 0/2 |
| BMI | 28.3 (6.1) | 28.1 | 27.2 (4.7) | 27.7 (5.7) | 28.9 (5.4) | 25.7, 51.2 (2 values) |
| Cre/µmol/L (SD) | 154.6 (79.2) | 91 | 98.3 (14.3) | 148.5 (38.2) | 237.0 (46.2) | 426, 434 (2 values) |
| eGFR/mL/min/per 1.73 m2 (SD) | 49.4 (20.8) | 91 | 69.9 (7.1) | 43.0 (8.9) | 23.7 (2.9) | 13, 14 (2 values) |
| Proteinuria level/g/ 24 h (SD) | 0.847 (1.965) | 0.126 | 0.286 (0.443) | 0.985 (2.081) | 0.6684 (0.788) | 1.17, 11.024 (2 values) |
| Parameter | CG | ACR | AMR |
|---|---|---|---|
| Sample size | 35 | 10 | 10 |
| Age/years (SD) | 56.6 (16.1) | 50.0 (12.7) | 46.0 (14.2) |
| Gender (F/M) | 14/21 | 4/6 | 5/5 |
| BMI | 28.6 (5.2) | 29.5 (8.3) | 26.1 (5.9) |
| Cre/µmol/L (SD) | 123 (43.2) | 185.5 (95.9) | 234.2 (89.3) |
| eGFR/mL/min/per 1.73 m2 (SD) | 58.0 (18.1) | 40.6 (18.0) | 28.3 (11.2) |
| Proteinuria level/g/24 h (SD) | 0.365 (0.458) | 1.222 (2.828) | 2.155 (3.076) |
| Cre (µmol/L) | eGFR (mL/min/per 1.73 m2) | Proteinuria (g/24 h) | ||||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| Cre (µmol/L) | 1 | - | −0.83 | 2.9 × 10−15 * | 0.47 | 0.00024 * |
| eGFR (mL/min/per 1.73 m2) | −0.83 | 2.9 × 10−15 * | 1 | - | −0.34 | 0.0089 * |
| Stage | 0.84 | 6.6 × 10−16 * | −0.92 | 1.6 × 10−23 * | 0.34 | 0.010 * |
| Proteinuria (g/24 h) | 0.47 | 0.00024 * | −0.34 | 0.0089 * | 1 | - |
| Alanine | 0.28 | 0.044 * | −0.11 | 0.40 | 0.02 | 0.86 |
| Acetate | 0.17 | 0.19 | −0.29 | 0.029 * | 0.06 | 0.64 |
| Citrate | 0.10 | 0.43 | −0.29 | 0.031 * | −0.04 | 0.76 |
| Phenylalanine | 0.45 | 0.00044 * | −0.46 | 0.00030 * | −0.06 | 0.62 |
| Tyrosine | −0.26 | 0.05 | 0.18 | 0.16 | −0.20 | 0.12 |
| Glutamine | 0.48 | 0.00016 * | −0.39 | 0.0026 * | 0 | 0.98 |
| Ketoleucine | −0.14 | 0.27 | 0.30 | 0.02 * | −0.36 | 0.0055 * |
| Ketoisoleucine | −0.15 | 0.26 | 0.23 | 0.08 | −0.28 | 0.033 * |
| Ketovaline | −0.20 | 0.12 | 0.26 | 0.05 | −0.43 | 0.0010 * |
| Creatinine | 0.98 | 0.00016 * | −0.83 | 3.4 × 10−15 * | 0.42 | 0.0013 * |
| Proline | 0.45 | 0.00049 * | −0.33 | 0.011 * | 0.02 | 0.84 |
| Histidine | 0.66 | 3.6 × 10−8 * | −0.39 | 0.0031 * | 0.13 | 0.32 |
| Metabolite | Stage 2, 3, 4 | Stage 2–3 | Stage 2–4 | Stage 3–4 |
|---|---|---|---|---|
| Glutamine | 0.035 * (2 < 3 < 4) | 0.155 (2 < 3) | 0.01 * (2 < 4) | 0.17 (3 < 4) |
| Phenylalanine | 0.0019 * (2 < 3 < 4) | 0.019 * (2 < 3) | 0.00072 * (2 < 4) | 0.14 (3 < 4) |
| Tyrosine | 0.033 * (2 > 3 > 4) | 0.34 (2 > 3) | 0.0091 * (2 > 4) | 0.06 (3 > 4) |
| Histidine | 0.085 (2 < 3 < 4) | 0.57 (2 < 3) | 0.028 * (2 < 4) | 0.09 (3 < 4) |
| Proline | 0.019 * (2 < 3 < 4) | 0.08 (2 < 3) | 0.0065 * (2 < 4) | 0.16 (3 < 4) |
| Ketoleucine | 0.028 * (2 > 3 > 4) | 0.04 * (2 > 3) | 0.015 * (2 > 4) | 0.43 (3 > 4) |
| Ketoisoleucine | 0.082 (2 > 3 > 4) | 0.39 (2 > 3) | 0.041 * (2 > 4) | 0.043 * (3 > 4) |
| Ketovaline | 0.142 (2 > 3 > 4) | 0.41 (2 > 3) | 0.047 * (2 > 4) | 0.17 (3 > 4) |
| Metabolite | CG-ACR-AMR | CG-ACR | CG-AMR | ACR-AMR |
|---|---|---|---|---|
| Lactate | 0.010 * | 0.31 | 0.0096 * (CG > AMR) | 0.0047 * (ACR > AMR) |
| Valine | 0.014 * | 0.0039 * (CG > ACR) | 0.35 | 0.10 |
| Leucine | 0.021 * | 0.0059 * (CG > ACR) | 0.36 | 0.12 |
| Isoleucine | 0.11 | 0.035 * (CG >ACR) | 0.64 | 0.17 |
| Tyrosine | 0.067 | 0.25 | 0.026 * (CG > AMR) | 0.41 |
| Glutamine | 0.062 | 0.041 * (CG < ACR) | 0.048 * (CG <AMR) | 0.99 |
| Creatinine | 0.00016 * | 0.0086 * (CG < ACR) | 0.00019 * (CG < AMR) | 0.44 |
| Proline | 0.015 * | 0.53 | 0.0039 * (CG < AMR) | 0.082 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranovicova, E.; Vnucak, M.; Granak, K.; Lehotsky, J.; Kadasova, N.; Miklusica, J.; Dedinska, I. Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients. Metabolites 2022, 12, 661. https://doi.org/10.3390/metabo12070661
Baranovicova E, Vnucak M, Granak K, Lehotsky J, Kadasova N, Miklusica J, Dedinska I. Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients. Metabolites. 2022; 12(7):661. https://doi.org/10.3390/metabo12070661
Chicago/Turabian StyleBaranovicova, Eva, Matej Vnucak, Karol Granak, Jan Lehotsky, Nina Kadasova, Juraj Miklusica, and Ivana Dedinska. 2022. "Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients" Metabolites 12, no. 7: 661. https://doi.org/10.3390/metabo12070661
APA StyleBaranovicova, E., Vnucak, M., Granak, K., Lehotsky, J., Kadasova, N., Miklusica, J., & Dedinska, I. (2022). Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients. Metabolites, 12(7), 661. https://doi.org/10.3390/metabo12070661