Microbial Virulence Factors, Antimicrobial Resistance Genes, Metabolites, and Synthetic Chemicals in Cabins of Commercial Aircraft
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Dust Sample Collection
2.2. Shotgun Metagenomic Sequencing
2.3. Dust Metabolites/Chemical Profiling via LC/MS
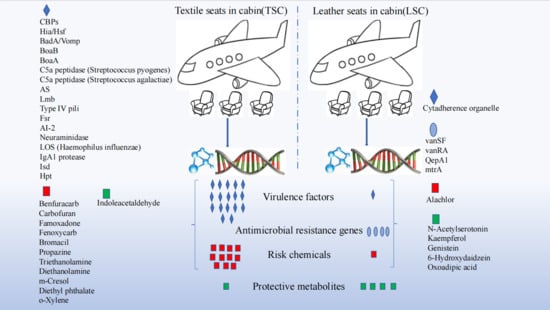
3. Results
3.1. Microorganisms and Functional Genes in TSC and LSC
3.2. Virulence Factors and Antibiotic Resistance Genes in TSC and LSC
3.3. Potential Risk and Protective Chemicals in TSC and LSC
3.4. Comparison of Exposure between Aircraft and a Common School Environment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilder-Smith, A.; Leong, H.N.; Villacian, J.S. In-flight transmission of Severe Acute Respiratory Syndrome (SARS): A case report. J. Travel Med. 2003, 10, 299–300. [Google Scholar] [CrossRef] [Green Version]
- Bylicki, O.; Paleiron, N.; Janvier, F. An Outbreak of Covid-19 on an Aircraft Carrier. New Engl. J. Med. 2021, 384, 976–977. [Google Scholar] [CrossRef]
- Bekö, G.; Allen, J.G.; Weschler, C.J.; Vallarino, J.; Spengler, J.D. Impact of cabin ozone concentrations on passenger reported symptoms in commercial aircraft. PLoS ONE 2015, 10, e0128454. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Lindgren, T.; Wieslander, G.; Janson, C.; Norbäck, D. Respiratory Illness and Allergy Related to Work and Home Environment among Commercial Pilots. PLoS ONE 2016, 11, e0164954. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Lindgren, T.; Norbäck, D. Medical symptoms among pilots associated with work and home environments: A 3-year cohort study. Aerosp. Med. Hum. Perform. 2015, 86, 458–465. [Google Scholar] [CrossRef]
- Sun, Y.; Fu, X.; Li, Y.; Yuan, Q.; Ou, Z.; Lindgren, T.; Deng, Y.; Norbäck, D. Shotgun Metagenomics Of Dust Microbiome From Flight Deck And Cabin In Civil Aviation Aircraft. Indoor Air 2020, 30, 1199–1212. [Google Scholar] [CrossRef]
- Fu, X.; Ou, Z.; Sun, Y. Indoor microbiome and allergic diseases: From theoretical advances to prevention strategies. Eco-Environ. Health. 2022, 1, 133–146. [Google Scholar] [CrossRef]
- Fu, X.; Lindgren, T.; Guo, M.; Cai, G.H.; Lundgren, H.; Norback, D. Furry pet allergens, fungal DNA and microbial volatile organic compounds (MVOCs) in the commercial aircraft cabin environment. Environ. Sci. Process. Impacts 2013, 15, 1228–1234. [Google Scholar] [CrossRef]
- Cross, A.S. What is a virulence factor? Crit. Care 2008, 12, 196. [Google Scholar] [CrossRef] [Green Version]
- Harbottle, H.; Thakur, S.; Zhao, S.; White, D.G. Genetics of antimicrobial resistance. Anim. Biotechnol. 2006, 17, 111–124. [Google Scholar] [CrossRef]
- Wang, Y.; Song, F.; Zhu, J.; Zhang, S.; Yang, Y.; Chen, T.; Tang, B.; Dong, L.; Ding, N.; Zhang, Q.; et al. GSA: Genome Sequence Archive. Genom. Proteom. Bioinform. 2017, 15, 14–18. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids. Res. 2010, 38, e132. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 1–18. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Morton, J.T.; Aksenov, A.A.; Nothias, L.F.; Foulds, J.R.; Quinn, R.A.; Badri, M.H.; Swenson, T.L.; Van Goethem, M.W.; Northen, T.R.; Vazquez-Baeza, Y.; et al. Learning representations of microbe–metabolite interactions. Nat. Methods 2019, 16, 1306–1314. [Google Scholar] [CrossRef] [Green Version]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, R.B. CPDB: Carcinogenic Potency Database. Med. Ref. Serv. Q. 2008, 27, 303–311. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.; Ou, Z.; Meng, Y.; Chen, Y.; Lin, R.; Hashim, J.H.; Hashim, Z.; Wieslander, G.; Chen, Q.; et al. Indoor microbiome, microbial and plant metabolites, chemical compounds and asthma symptoms in junior high school students: A multicentre association study in Malaysia. Eur. Respir. J. 2022, 60, 2200260. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Zhu, S.; Tonnessen, T.I. Ethyl pyruvate is a novel anti-inflammatory agent to treat multiple inflammatory organ injuries. J. Inflamm. 2016, 13, 37. [Google Scholar] [CrossRef] [Green Version]
- Lowes, D.A.; Almawash, A.M.; Webster, N.R.; Reid, V.L.; Galley, H.F. Melatonin and structurally similar compounds have differing effects on inflammation and mitochondrial function in endothelial cells under conditions mimicking sepsis. Br. J. Anaesth. 2011, 107, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhao, Y.; Qi, D.; He, J.; Wang, D. Tangeretin attenuates lipopolysaccharide-induced acute lung injury through Notch signaling pathway via suppressing Th17 cell response in mice. Microb. Pathog. 2020, 138, 103826. [Google Scholar] [CrossRef]
- Fu, X.; Ou, Z.; Zhang, M.; Meng, Y.; Li, Y.; Wen, J.; Hu, Q.; Zhang, X.; Norbäck, D.; Deng, Y.; et al. Indoor bacterial, fungal and viral species and functional genes in urban and rural schools in Shanxi Province, China–association with asthma, rhinitis and rhinoconjunctivitis in high school students. Microbiome 2021, 9, 138. [Google Scholar] [CrossRef]
- Valo, E.; Colombo, M.; Sandholm, N.; McGurnaghan, S.J.; Blackbourn, L.A.K.; Dunger, D.B.; McKeigue, P.M.; Forsblom, C.; Groop, P.-H.; Colhoun, H.M.; et al. Effect of serum sample storage temperature on metabolomic and proteomic biomarkers. Sci. Rep. 2022, 12, 4571. [Google Scholar] [CrossRef]
- Gosink, K.K.; Mann, E.R.; Guglielmo, C.; Tuomanen, E.I.; Masure, H.R. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 2000, 68, 5690–5695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St Geme, J.W., 3rd; Cutter, D.; Barenkamp, S.J. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 1996, 178, 6281–6287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaglenov, K. Survival and Transmission of Selected Pathogens on Airplane Cabin Surfaces and Selection of Phages Specific for Campylobacter jejuni. Auburn Univ. 2014. Available online: https://etd.auburn.edu/handle/10415/4066?show=full (accessed on 28 April 2014).
- Leung, M.H.Y.; Tong, X.; Bøifot, K.O.; Bezdan, D.; Butler, D.J.; Danko, D.C.; Gohli, J.; Green, D.C.; Hernandez, M.T.; Kelly, F.J.; et al. Characterization of the public transit air microbiome and resistome reveals geographical specificity. Microbiome 2021, 9, 112. [Google Scholar] [CrossRef]
- Lax, S.; Sangwan, N.; Smith, D.; Larsen, P.; Handley, K.M.; Richardson, M.; Guyton, K.; Krezalek, M.; Shogan, B.D.; Defazio, J.; et al. Bacterial colonization and succession in a newly opened hospital. Sci. Transl. Med. 2017, 9, eaah6500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chng, K.R.; Li, C.; Bertrand, D.; Ng, A.H.Q.; Kwah, J.S.; Low, H.M.; Tong, C.; Natrajan, M.; Zhang, M.H.; Xu, L.; et al. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 2020, 26, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.T.; Caldas, R.; Pereira, T. Allergic contact dermatitis caused by triethanolamine in an ultrasound gel. Contact Dermat. 2020, 82, 64–65. [Google Scholar] [CrossRef] [PubMed]
- Mertens, S.; Gilissen, L.; Goossens, A. Allergic contact dermatitis caused by cocamide diethanolamine. Contact Dermat. 2016, 75, 20–24. [Google Scholar] [CrossRef]
- Weaver, J.A.; Beverly, B.E.J.; Keshava, N.; Mudipalli, A.; Arzuaga, X.; Cai, C.; Hotchkiss, A.K.; Makris, S.L.; Yost, E.E. Hazards of diethyl phthalate (DEP) exposure: A systematic review of animal toxicology studies. Environ. Int. 2020, 145, 105848. [Google Scholar] [CrossRef]
- Gamer, A.O.; Rossbacher, R.; Kaufmann, W.; van Ravenzwaay, B. The inhalation toxicity of di- and triethanolamine upon repeated exposure. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2008, 46, 2173–2183. [Google Scholar] [CrossRef]
- Tunctan, B.; Ozveren, E.; Korkmaz, B.; Buharalioglu, C.K.; Tamer, L.; Degirmenci, U.; Atik, U. Nitric oxide reverses endotoxin-induced inflammatory hyperalgesia via inhibition of prostacyclin production in mice. Pharmacol. Res. 2006, 53, 177–192. [Google Scholar] [CrossRef]
- Devi, K.P.; Malar, D.S.; Nabavi, S.F.; Sureda, A.; Xiao, J.; Nabavi, S.M.; Daglia, M. Kaempferol and inflammation: From chemistry to medicine. Pharmacol. Res. 2015, 99, 1–10. [Google Scholar] [CrossRef]
- Han, X.; Sun, S.; Sun, Y.; Song, Q.; Zhu, J.; Song, N.; Chen, M.; Sun, T.; Xia, M.; Ding, J.; et al. Small molecule-driven NLRP3 inflammation inhibition via interplay between ubiquitination and autophagy: Implications for Parkinson disease. Autophagy 2019, 15, 1860–1881. [Google Scholar] [CrossRef]
- Kim, E.; Kang, Y.-G.; Kim, J.H.; Kim, Y.-J.; Lee, T.R.; Lee, J.; Kim, D.; Cho, J.Y. The Antioxidant and Anti-Inflammatory Activities of 8-Hydroxydaidzein (8-HD) in Activated Macrophage-Like RAW264.7 Cells. Int J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Zhang, M.; Yuan, Y.; Chen, Y.; Ou, Z.; Hashim, Z.; Hashim, J.H.; Zhang, X.; Zhao, Z.; Norbäck, D.; et al. Microbial Virulence Factors, Antimicrobial Resistance Genes, Metabolites, and Synthetic Chemicals in Cabins of Commercial Aircraft. Metabolites 2023, 13, 343. https://doi.org/10.3390/metabo13030343
Fu X, Zhang M, Yuan Y, Chen Y, Ou Z, Hashim Z, Hashim JH, Zhang X, Zhao Z, Norbäck D, et al. Microbial Virulence Factors, Antimicrobial Resistance Genes, Metabolites, and Synthetic Chemicals in Cabins of Commercial Aircraft. Metabolites. 2023; 13(3):343. https://doi.org/10.3390/metabo13030343
Chicago/Turabian StyleFu, Xi, Mei Zhang, Yiwen Yuan, Yang Chen, Zheyuan Ou, Zailina Hashim, Jamal Hisham Hashim, Xin Zhang, Zhuohui Zhao, Dan Norbäck, and et al. 2023. "Microbial Virulence Factors, Antimicrobial Resistance Genes, Metabolites, and Synthetic Chemicals in Cabins of Commercial Aircraft" Metabolites 13, no. 3: 343. https://doi.org/10.3390/metabo13030343
APA StyleFu, X., Zhang, M., Yuan, Y., Chen, Y., Ou, Z., Hashim, Z., Hashim, J. H., Zhang, X., Zhao, Z., Norbäck, D., & Sun, Y. (2023). Microbial Virulence Factors, Antimicrobial Resistance Genes, Metabolites, and Synthetic Chemicals in Cabins of Commercial Aircraft. Metabolites, 13(3), 343. https://doi.org/10.3390/metabo13030343