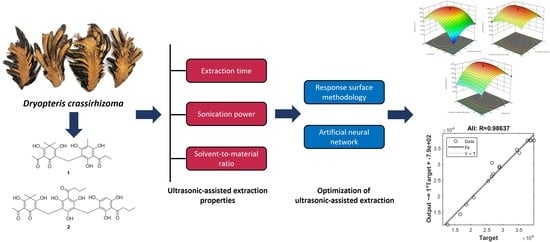
Optimization of Ultrasonic-Assisted Extraction of α-Glucosidase Inhibitors from Dryopteris crassirhizoma Using Artificial Neural Network and Response Surface Methodology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Procedures
2.2. Plant Material
2.3. Extraction and Isolation
2.4. Evaluation of the Inhibitory Activity against α-Glucosidase
2.5. HPLC Analysis
2.6. Selection of Variables
2.7. Box–Behnken Design for Optimization
2.8. ANN
3. Results and Discussion
3.1. Identification of Isolated Compounds from D. crassirhizoma
3.2. Inhibitory Activity against α-Glucosidase
3.3. Development of the HPLC Analysis Method
3.4. RSM Optimization
− 1.515625 × AB + 3.4375 × AC + 0.576719 × BC
− 15.430625 × A2 − 1.691836 × B2 − 1.914219 × C2
3.5. Combined Effect of Solvent Concentration, Power, and Extraction Time
3.6. ANN
3.7. Validation of the Optimal Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Z.L.; Leng, C.H.; Wang, Z.T. Identification of Dryopteris crassirhizoma and the adulterant species based on cpDNA rbcL and translated amino acid sequences. Planta Med. 2007, 73, 1230–1233. [Google Scholar] [CrossRef]
- Lee, J.; Nho, Y.H.; Yun, S.K.; Hwang, Y.S. Anti-invasive and anti-tumor effects of Dryopteris crassirhizoma extract by disturbing actin polymerization. Integr. Cancer Ther. 2019, 18, 1534735419851197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Hu, W.; Zhang, H.; Ding, C.; Huang, Y.; Liao, J.; Zhang, Z.; Yuan, S.; Chen, Y.; Yuan, M. Antioxidant and immunomodulatory activities of polysaccharides from the rhizome of Dryopteris crassirhizoma Nakai. Int. J. Biol. Macromol. 2019, 130, 238–244. [Google Scholar] [CrossRef]
- Piao, C.H.; Bui, T.T.; Fan, Y.J.; Nguyen, T.V.; Shin, D.U.; Song, C.H.; Lee, S.Y.; Shin, H.S.; Kim, H.T.; Chai, O.H. In vivo and in vitro anti-allergic and anti-inflammatory effects of Dryopteris crassirhizoma through the modulation of the NF-ĸB signaling pathway in an ovalbumin-induced allergic asthma mouse model. Mol. Med. Rep. 2020, 22, 3597–3606. [Google Scholar] [CrossRef] [PubMed]
- Phong, N.V.; Oanh, V.T.; Yang, S.Y.; Choi, J.S.; Min, B.S.; Kim, J.A. PTP1B inhibition studies of biological active phloroglucinols from the rhizomes of Dryopteris crassirhizoma: Kinetic properties and molecular docking simulation. Int. J. Biol. Macromol. 2021, 188, 719–728. [Google Scholar] [CrossRef]
- Phong, N.V.; Zhao, Y.; Min, B.S.; Yang, S.Y.; Kim, J.A. Inhibitory activity of bioactive phloroglucinols from the rhizomes of Dryopteris crassirhizoma on Escherichia coli β-glucuronidase: Kinetic analysis and molecular docking studies. Metabolites 2022, 12, 938. [Google Scholar] [CrossRef]
- Yim, N.-H.; Lee, J.-J.; Lee, B.; Li, W.; Ma, J.Y. Antiplatelet activity of acylphloroglucinol derivatives isolated from Dryopteris crassirhizoma. Molecules 2019, 24, 2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, P.; Rao, K.; Jiang, N.; Liu, D.; Wang, R.; Gao, Y.; Liu, X.; Deng, S.; Zhou, Y.; Chen, X.; et al. Structure elucidation, biogenesis, and bioactivities of acylphloroglucinol-derived meroterpenoid enantiomers from Dryopteris crassirhizoma. Bioorg. Chem. 2022, 119, 105567. [Google Scholar] [CrossRef]
- Jin, Y.-H.; Jeon, S.; Lee, J.; Kim, S.; Jang, M.S.; Park, C.M.; Song, J.H.; Kim, H.R.; Kwon, S. Anticoronaviral activity of the natural phloroglucinols, dryocrassin ABBA and filixic acid ABA from the rhizome of Dryopteris crassirhizoma by targeting the main protease of SARS-CoV-2. Pharmaceutics 2022, 14, 376. [Google Scholar] [CrossRef]
- Pham, V.C.; Kim, O.; Lee, J.-H.; Min, B.S.; Kim, J.A. Inhibitory effects of phloroglucinols from the roots of Dryopteris crassirhizoma on melanogenesis. Phytochem. Lett. 2017, 21, 51–56. [Google Scholar] [CrossRef]
- Wang, J.; Yan, Y.-T.; Fu, S.-Z.; Peng, B.; Bao, L.-L.; Zhang, Y.-L.; Hu, J.-H.; Zeng, Z.-P.; Geng, D.-H.; Gao, Z.-P. Anti-influenza virus (H5N1) activity screening on the phloroglucinols from rhizomes of Dryopteris crassirhizoma. Molecules 2017, 22, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuk, H.J.; Kim, J.-Y.; Sung, Y.-Y.; Kim, D.-S. Phloroglucinol derivatives from Dryopteris crassirhizoma as potent xanthine oxidase inhibitors. Molecules 2021, 26, 122. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Le Ba, V.; Rustam, R.; Cho, C.W.; Yang, S.Y.; Su, X.D.; Kim, Y.H.; Kang, J.S. Isolation of bioactive components with soluble epoxide hydrolase inhibitory activity from Stachys sieboldii MiQ. by ultrasonic-assisted extraction optimized using response surface methodology. Prep. Biochem. Biotechnol. 2021, 51, 395–404. [Google Scholar] [CrossRef]
- Vinatoru, M. Ultrasonically assisted extraction (UAE) of natural products some guidelines for good practice and reporting. Ultrason. Sonochem. 2015, 25, 94–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lianfu, Z.; Zelong, L. Optimization and comparison of ultrasound/microwave assisted extraction (UMAE) and ultrasonic assisted extraction (UAE) of lycopene from tomatoes. Ultrason. Sonochem. 2008, 15, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Srivastav, S.; Sharanagat, V.S. Ultrasound assisted extraction (UAE) of bioactive compounds from fruit and vegetable processing by-products: A review. Ultrason. Sonochem. 2021, 70, 105325. [Google Scholar] [CrossRef]
- Huang, S.-M.; Kuo, C.-H.; Chen, C.-A.; Liu, Y.-C.; Shieh, C.-J. RSM and ANN modeling-based optimization approach for the development of ultrasound-assisted liposome encapsulation of piceid. Ultrason. Sonochem. 2017, 36, 112–122. [Google Scholar] [CrossRef]
- Maran, J.P.; Priya, B. Comparison of response surface methodology and artificial neural network approach towards efficient ultrasound-assisted biodiesel production from muskmelon oil. Ultrason. Sonochem. 2015, 23, 192–200. [Google Scholar] [CrossRef]
- Suryawanshi, N.; Naik, S.; Eswari, J.S. Extraction and optimization of exopolysaccharide from Lactobacillus sp. using response surface methodology and artificial neural networks. Prep. Biochem. Biotechnol. 2019, 49, 987–996. [Google Scholar] [CrossRef]
- Phong, N.V.; Yang, S.Y.; Min, B.S.; Kim, J.A. Insights into the inhibitory activity and mechanism of action of flavonoids from the stems and branches of Acer mono Maxim. against α-glucosidase via kinetic analysis, molecular docking, and molecular dynamics simulations. J. Mol. Struct. 2023, 1282, 135188. [Google Scholar] [CrossRef]
- Martin, A.E.; Montgomery, P.A. Acarbose: An α-glucosidase inhibitor. Am. J. Health-Syst. Pharm. 1996, 53, 2277–2290. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Kim, J.H.; Jang, H.D.; Yang, S.Y.; Kim, Y.H. Inhibitory activity of minor phlorotannins from Ecklonia cava on α-glucosidase. Food Chem. 2018, 257, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Starzec, A.; Włodarczyk, M.; Kunachowicz, D.; Dryś, A.; Kepinska, M.; Fecka, I. Polyphenol profile of Cistus × incanus L. and its relevance to antioxidant effect and α-glucosidase inhibition. Antioxidants 2023, 12, 553. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Cho, C.W.; Lee, J.I.; Vinh, L.B.; Kim, K.T.; Cho, I.S. An investigation of the inhibitory mechanism of α-glucosidase by chysalodin from Aloe vera. Int. J. Biol. Macromol. 2020, 147, 314–318. [Google Scholar] [CrossRef]
- Yi, S.; Su, Y.; Qi, B.; Su, Z.; Wan, Y. Application of response surface methodology and central composite rotatable design in optimizing the preparation conditions of vinyltriethoxysilane modified silicalite/polydimethylsiloxane hybrid pervaporation membranes. Sep. Purif. Technol. 2010, 71, 252–262. [Google Scholar] [CrossRef]
- Saleh, B.; Ma, A.; Fathi, R.; Radhika, N.; Yang, G.; Jiang, J. Optimized mechanical properties of magnesium matrix composites using RSM and ANN. Mater. Sci. Eng. B 2023, 290, 116303. [Google Scholar] [CrossRef]
- Said, F.M.; Gan, J.Y.; Sulaiman, J. Correlation between response surface methodology and artificial neural network in the prediction of bioactive compounds of unripe Musa acuminata peel. Eng. Sci. Technol. Int. J. 2020, 23, 781–787. [Google Scholar] [CrossRef]





| Extract or Fraction | 100 µg (%) | IC50 (µg/mL) | Compound | 100 µM (%) | IC50 (µM) |
|---|---|---|---|---|---|
| Ethanol extract | 73.8 ± 0.19 | 34.8 ± 1.43 | 1 | <50 | – |
| CHCl3 fraction | 71.6 ± 1.39 | 25.8 ± 0.68 | 2 | 59.5 ± 1.00 | 34.0 ± 0.14 |
| EtOAc fraction | 77.7 ± 0.44 | 15.2 ± 0.57 | |||
| Aqueous fraction | 79.9 ± 1.71 | 22.2 ± 1.92 | Acarbose a | 21.7 ± 1.43 | 329.2 ± 0.35 |
| Run | A: Sonication Time (min) | B: Sonication Power (W) | C: Solvent-to-Material Ratio (mL/g) | Response Peak Area Y (AU/min, ×104) |
|---|---|---|---|---|
| 1 | 80 (−1) | 240 (−1) | 80 (0) | 12,800 |
| 2 | 100 (0) | 400 (+1) | 40 (−1) | 28,960 |
| 3 | 120 (+1) | 240 (−1) | 80 (0) | 20,920 |
| 4 | 80 (−1) | 320 (0) | 40 (−1) | 28,960 |
| 5 | 100 (0) | 320 (0) | 80 (0) | 38,780 |
| 6 | 120 (+1) | 400 (+1) | 80 (0) | 25,440 |
| 7 | 100 (0) | 320 (0) | 80 (0) | 38,665 |
| 8 | 100 (0) | 320 (0) | 80 (0) | 37,760 |
| 9 | 120 (+1) | 320 (0) | 120 (+1) | 35,160 |
| 10 | 80 (−1) | 320 (0) | 120 (+1) | 26,520 |
| 11 | 100 (0) | 240 (−1) | 120 (+1) | 16,658 |
| 12 | 120 (+1) | 320 (0) | 40 (−1) | 26,600 |
| 13 | 80 (−1) | 400 (+1) | 80 (0) | 27,020 |
| 14 | 100 (0) | 320 (0) | 80 (0) | 37,760 |
| 15 | 100 (0) | 320 (0) | 80 (0) | 39,760 |
| 16 | 100 (0) | 400 (+1) | 120 (+1) | 34,400 |
| 17 | 100 (0) | 240 (−1) | 40 (−1) | 18,600 |
| Source | Sum of Squares | df 1 | Mean Square | F-Value 2 | p-Value | Remarks |
|---|---|---|---|---|---|---|
| Model | 1.127 × 109 | 9 | 1.253 × 108 | 60.05 | <0.0001 | significant |
| A–Sonication time | 2.054 × 107 | 1 | 2.054 × 107 | 9.85 | 0.0164 | |
| B–Power | 2.743 × 108 | 1 | 2.743 × 108 | 131.48 | <0.0001 | |
| C–Solvent-to-material-ratio | 1.156 × 107 | 1 | 1.156 × 107 | 5.54 | 0.0508 | |
| AB | 2.352 × 107 | 1 | 2.352 × 107 | 11.28 | 0.0121 | |
| AC | 3.025 × 107 | 1 | 3.025 × 107 | 14.50 | 0.0066 | |
| BC | 1.362 × 107 | 1 | 1.362 × 107 | 6.53 | 0.0378 | |
| A² | 1.604 × 108 | 1 | 1.604 × 108 | 76.90 | <0.0001 | |
| B² | 4.936 × 108 | 1 | 4.936 × 108 | 236.65 | <0.0001 | |
| C² | 3.950 × 107 | 1 | 3.950 × 107 | 18.93 | 0.0033 | |
| Residual | 1.460 × 107 | 7 | 2.086 × 106 | |||
| Lack of Fit | 1.182 × 107 | 3 | 3.941 × 106 | 5.67 | 0.0634 | not significant |
| Pure Error | 2.778 × 106 | 4 | 6.946 × 105 | |||
| Cor Total 3 | 1.142 × 109 | 16 | ||||
| R2 | 0.9872 | Adjusted R2 | 0.9969 | |||
| C.V. % | 4.9600 | Predicted R2 | 0.9853 |
| Run | Sonication Time (min) | Sonication Power (W) | Solvent-to-Material Ratio (mL/g) | Actual Value | Predicted (RSM) | Predicted (ANN) |
|---|---|---|---|---|---|---|
| 1 | 80 | 240 | 80 | 12,800 | 11,662 | 11,164 |
| 2 | 100 | 400 | 40 | 28,960 | 27,462 | 29,371 |
| 3 | 120 | 240 | 80 | 20,920 | 19,717 | 19,638 |
| 4 | 80 | 320 | 40 | 28,960 | 29,255 | 29,008 |
| 5 | 100 | 320 | 80 | 38,780 | 38,545 | 37,478 |
| 6 | 120 | 400 | 80 | 25,440 | 26,578 | 24,788 |
| 7 | 100 | 320 | 80 | 38,665 | 38,545 | 37,478 |
| 8 | 100 | 320 | 80 | 37,760 | 38,545 | 37,478 |
| 9 | 120 | 320 | 120 | 35,160 | 34,865 | 33,546 |
| 10 | 80 | 320 | 120 | 26,520 | 26,160 | 26,283 |
| 11 | 100 | 240 | 120 | 16,658 | 18,156 | 14,389 |
| 12 | 120 | 320 | 40 | 26,600 | 26,960 | 30,494 |
| 13 | 80 | 400 | 80 | 27,020 | 28,223 | 27,084 |
| 14 | 100 | 320 | 80 | 37,760 | 38,545 | 37,478 |
| 15 | 100 | 320 | 80 | 39,760 | 38,545 | 37,478 |
| 16 | 100 | 400 | 120 | 34,400 | 33,558 | 34,557 |
| 17 | 100 | 240 | 40 | 18,600 | 19,443 | 17,434 |
| A (min) | B (W) | C (mL/g) | Y (AU/min, ×104) | |
|---|---|---|---|---|
| Predicted (RSM) | 103.03 | 342.69 | 94.00 | 39,706.2 |
| Predicted (ANN) | 103.0 | 340.0 | 94.0 | 39,562.1 |
| Experimental | 103.0 | 340.0 | 94.0 | 38,575.2 |
| Matching (RSM, %) | 97.15% | |||
| Matching (ANN, %) | 97.51% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phong, N.V.; Gao, D.; Kim, J.A.; Yang, S.Y. Optimization of Ultrasonic-Assisted Extraction of α-Glucosidase Inhibitors from Dryopteris crassirhizoma Using Artificial Neural Network and Response Surface Methodology. Metabolites 2023, 13, 557. https://doi.org/10.3390/metabo13040557
Phong NV, Gao D, Kim JA, Yang SY. Optimization of Ultrasonic-Assisted Extraction of α-Glucosidase Inhibitors from Dryopteris crassirhizoma Using Artificial Neural Network and Response Surface Methodology. Metabolites. 2023; 13(4):557. https://doi.org/10.3390/metabo13040557
Chicago/Turabian StylePhong, Nguyen Viet, Dan Gao, Jeong Ah Kim, and Seo Young Yang. 2023. "Optimization of Ultrasonic-Assisted Extraction of α-Glucosidase Inhibitors from Dryopteris crassirhizoma Using Artificial Neural Network and Response Surface Methodology" Metabolites 13, no. 4: 557. https://doi.org/10.3390/metabo13040557
APA StylePhong, N. V., Gao, D., Kim, J. A., & Yang, S. Y. (2023). Optimization of Ultrasonic-Assisted Extraction of α-Glucosidase Inhibitors from Dryopteris crassirhizoma Using Artificial Neural Network and Response Surface Methodology. Metabolites, 13(4), 557. https://doi.org/10.3390/metabo13040557