Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review
Abstract
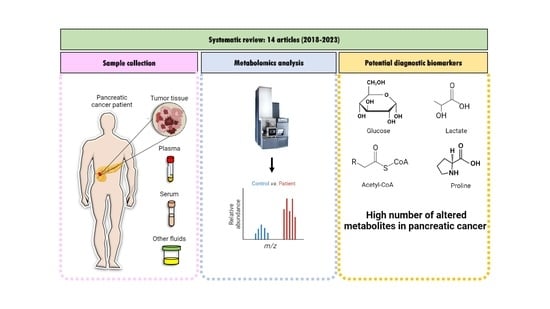
:1. Introduction
2. Materials and Methods
2.1. Research Question
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Data Sources
2.5. Study Selection
2.6. Data Extraction
3. Results
4. Discussion
4.1. Metabolites in Tissue
4.2. Metabolites in Serum
4.3. Metabolites in Plasma
4.4. Metabolites in Other Fluids
4.5. Metabolic Pathways
4.5.1. Amino Acid Metabolism
4.5.2. Carbohydrate Metabolism
4.5.3. Lipid Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol. Cancer Res. Treat. 2020, 19, 1533033820962117. [Google Scholar] [CrossRef] [PubMed]
- Lucenteforte, E.; La Vecchia, C.; Silverman, D.; Petersen, G.M.; Bracci, P.M.; Ji, B.T.; Bosetti, C.; Li, D.; Gallinger, S.; Miller, A.B.; et al. Alcohol Consumption and Pancreatic Cancer: A Pooled Analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann. Oncol. 2012, 23, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Bosetti, C.; Lucenteforte, E.; Silverman, D.T.; Petersen, G.; Bracci, P.M.; Ji, B.T.; Negri, E.; Li, D.; Risch, H.A.; Olson, S.H.; et al. Cigarette Smoking and Pancreatic Cancer: An Analysis from the International Pancreatic Cancer Case-Control Consortium (Panc4). Ann. Oncol. 2012, 23, 1880–1888. [Google Scholar] [CrossRef]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and Cancer Risk: Emerging Biological Mechanisms and Perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef]
- Brune, K.A.; Lau, B.; Palmisano, E.; Canto, M.; Goggins, M.G.; Hruban, R.H.; Klein, A.P. Importance of Age of Onset in Pancreatic Cancer Kindreds. J. Natl. Cancer Inst. 2010, 102, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.P. Pancreatic Cancer Epidemiology: Understanding the Role of Lifestyle and Inherited Risk Factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Amini, N.; Spolverato, G.; Gupta, R.; Margonis, G.A.; Kim, Y.; Wagner, D.; Rezaee, N.; Weiss, M.J.; Wolfgang, C.L.; Makary, M.M.; et al. Impact Total Psoas Volume on Short- and Long-Term Outcomes in Patients Undergoing Curative Resection for Pancreatic Adenocarcinoma: A New Tool to Assess Sarcopenia. J. Gastrointest. Surg. 2015, 19, 1593–1602. [Google Scholar] [CrossRef]
- Kim, J.-E.; Lee, K.T.; Lee, J.K.; Paik, S.W.; Rhee, J.C.; Choi, K.W. Clinical Usefulness of Carbohydrate Antigen 19-9 as a Screening Test for Pancreatic Cancer in an Asymptomatic Population. J. Gastroenterol. Hepatol. 2004, 19, 182–186. [Google Scholar] [CrossRef]
- Haab, B.B.; Huang, Y.; Balasenthil, S.; Partyka, K.; Tang, H.; Anderson, M.; Allen, P.; Sasson, A.; Zeh, H.; Kaul, K.; et al. Definitive Characterization of CA 19-9 in Resectable Pancreatic Cancer Using a Reference Set of Serum and Plasma Specimens. PLoS ONE 2015, 10, e0139049. [Google Scholar] [CrossRef] [Green Version]
- Wood, L.D.; Canto, M.I.; Jaffee, E.M.; Simeone, D.M. Pancreatic Cancer: Pathogenesis, Screening, Diagnosis, and Treatment. Gastroenterology 2022, 163, 386–402.e1. [Google Scholar] [CrossRef]
- Zhao, R.; Ren, S.; Li, C.; Guo, K.; Lu, Z.; Tian, L.; He, J.; Zhang, K.; Cao, Y.; Liu, S.; et al. Biomarkers for Pancreatic Cancer Based on Tissue and Serum Metabolomics Analysis in a Multicenter Study. Cancer Med. 2023, 12, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef]
- Keane, M.G.; Afghani, E. A Review of the Diagnosis and Management of Premalignant Pancreatic Cystic Lesions. J. Clin. Med. 2021, 10, 1284. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Liu, X.; Suriawinata, A.A. Pancreatic Ductal Adenocarcinoma and Its Precursor Lesions: Histopathology, Cytopathology, and Molecular Pathology. Am. J. Pathol. 2019, 189, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radon, T.P.; Massat, N.J.; Jones, R.; Alrawashdeh, W.; Dumartin, L.; Ennis, D.; Duffy, S.W.; Kocher, H.M.; Pereira, S.P.; Guarner (posthumous), L.; et al. Identification of a Three-Biomarker Panel in Urine for Early Detection of Pancreatic Adenocarcinoma. Clin. Cancer Res. 2015, 21, 3512–3521. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary Electrophoresis Mass Spectrometry-Based Saliva Metabolomics Identified Oral, Breast and Pancreatic Cancer-Specific Profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Rosty, C.; Goggins, M. Early Detection of Pancreatic Carcinoma. Hematol. Oncol. Clin. N. Am. 2002, 16, 37–52. [Google Scholar] [CrossRef]
- Mahajan, U.M.; Oehrle, B.; Sirtl, S.; Alnatsha, A.; Goni, E.; Regel, I.; Beyer, G.; Vornhülz, M.; Vielhauer, J.; Chromik, A.; et al. Independent Validation and Assay Standardization of Improved Metabolic Biomarker Signature to Differentiate Pancreatic Ductal Adenocarcinoma from Chronic Pancreatitis. Gastroenterology 2022, 163, 1407–1422. [Google Scholar] [CrossRef]
- Long, N.P.; Yoon, S.J.; Anh, N.H.; Nghi, T.D.; Lim, D.K.; Hong, Y.J.; Hong, S.-S.; Kwon, S.W. A Systematic Review on Metabolomics-Based Diagnostic Biomarker Discovery and Validation in Pancreatic Cancer. Metabolomics 2018, 14, 109. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Br. Med. J. 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Wanden-Berghe, C.; Sanz-Valero, J. Systematic Reviews in Nutrition: Standardized Methodology. Br. J. Nutr. 2012, 107, S3–S7. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wen, S.; Guo, P.; Liu, H.; Feng, J.; Huang, H. Understanding Metabolomic Characteristics of Pancreatic Ductal Adenocarcinoma by HR-MAS NMR Detection of Pancreatic Tissues. J. Pharm. Biomed. Anal. 2020, 190, 113546. [Google Scholar] [CrossRef] [PubMed]
- Unger, K.; Mehta, K.Y.; Kaur, P.; Wang, Y.; Menon, S.S.; Jain, S.K.; Moonjelly, R.A.; Suman, S.; Datta, K.; Singh, R.; et al. Metabolomics Based Predictive Classifier for Early Detection of Pancreatic Ductal Adenocarcinoma. Oncotarget 2018, 9, 23078–23090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Hu, Z.; Rong, S.; Xie, G.; Nie, G.; Liu, X.; Jin, G. Integrative Analysis of Metabolome and Gut Microbiota in Patients with Pancreatic Ductal Adenocarcinoma. J. Cancer 2022, 13, 1555–1564. [Google Scholar] [CrossRef]
- Wolrab, D.; Jirásko, R.; Cífková, E.; Höring, M.; Mei, D.; Chocholoušková, M.; Peterka, O.; Idkowiak, J.; Hrnčiarová, T.; Kuchař, L.; et al. Lipidomic Profiling of Human Serum Enables Detection of Pancreatic Cancer. Nat. Commun. 2022, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Michálková, L.; Horník, Š.; Sýkora, J.; Habartová, L.; Setnička, V.; Bunganič, B. Early Detection of Pancreatic Cancer in Type 2 Diabetes Mellitus Patients Based on 1H NMR Metabolomics. J. Proteome Res. 2021, 20, 1744–1753. [Google Scholar] [CrossRef]
- Morgell, A.; Reisz, J.A.; Ateeb, Z.; Davanian, H.; Reinsbach, S.E.; Halimi, A.; Gaiser, R.; Valente, R.; Arnelo, U.; Del Chiaro, M.; et al. Metabolic Characterization of Plasma and Cyst Fluid from Cystic Precursors to Pancreatic Cancer Patients Reveal Metabolic Signatures of Bacterial Infection. J. Proteome Res. 2021, 20, 2725–2738. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, L.; Kang, H.; Liu, J.; Zhang, J.; Zhao, J.; Liu, S. Metabolomics Identifies Biomarker Signatures to Differentiate Pancreatic Cancer from Type 2 Diabetes Mellitus in Early Diagnosis. Int. J. Endocrinol. 2021, 2021, 9990768. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Bantis, L.E.; Capello, M.; Scelo, G.; Dennison, J.B.; Patel, N.; Murage, E.; Vykoukal, J.; Kundnani, D.L.; Foretova, L.; et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J. Natl. Cancer Inst. 2019, 111, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Moore, H.B.; Culp-Hill, R.; Reisz, J.A.; Lawson, P.J.; Sauaia, A.; Schulick, R.D.; Del Chiaro, M.; Nydam, T.L.; Moore, E.E.; Hansen, K.C.; et al. The Metabolic Time Line of Pancreatic Cancer: Opportunities to Improve Early Detection of Adenocarcinoma. Am. J. Surg. 2019, 218, 1206–1212. [Google Scholar] [CrossRef]
- Tumas, J.; Baskirova, I.; Petrenas, T.; Norkuniene, J.; Strupas, K.; Sileikis, A. Towards a Personalized Approach in Pancreatic Cancer Diagnostics Through Plasma Amino Acid Analysis. Anticancer Res. 2019, 39, 2035–2042. [Google Scholar] [CrossRef]
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; González Maldonado, S.; Pilarsky, C.; Heidecke, C.-D.; Schatz, P.; et al. Metabolic Biomarker Signature to Differentiate Pancreatic Ductal Adenocarcinoma from Chronic Pancreatitis. Gut 2018, 67, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Ikeura, T.; Hori, Y.; Mitsuyama, T.; Miyoshi, H.; Shimatani, M.; Uchida, K.; Takaoka, M.; Ota, U.; Kamiya, A.; Takahashi, K.; et al. Effectiveness of Photodynamic Screening Using 5-Aminolevulinic Acid for the Diagnosis of Pancreatic Cancer. Anticancer Res. 2020, 40, 3571–3577. [Google Scholar] [CrossRef]
- Asai, Y.; Itoi, T.; Sugimoto, M.; Sofuni, A.; Tsuchiya, T.; Tanaka, R.; Tonozuka, R.; Honjo, M.; Mukai, S.; Fujita, M.; et al. Elevated Polyamines in Saliva of Pancreatic Cancer. Cancers 2018, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- San-Millán, I.; Brooks, G.A. Reexamining Cancer Metabolism: Lactate Production for Carcinogenesis Could Be the Purpose and Explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [Green Version]
- Vlachostergios, P.J.; Oikonomou, K.G.; Gibilaro, E.; Apergis, G. Elevated Lactic Acid Is a Negative Prognostic Factor in Metastatic Lung Cancer. Cancer Biomark. 2015, 15, 725–734. [Google Scholar] [CrossRef]
- Nava, G.M.; Madrigal Perez, L.A. Metabolic Profile of the Warburg Effect as a Tool for Molecular Prognosis and Diagnosis of Cancer. Expert. Rev. Mol. Diagn. 2022, 22, 439–447. [Google Scholar] [CrossRef]
- Pascual, G.; Domínguez, D.; Elosúa-Bayes, M.; Beckedorff, F.; Laudanna, C.; Bigas, C.; Douillet, D.; Greco, C.; Symeonidi, A.; Hernández, I.; et al. Dietary Palmitic Acid Promotes a Prometastatic Memory via Schwann Cells. Nature 2021, 599, 485–490. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. Lipids in Cancer: A Global View of the Contribution of Lipid Pathways to Metastatic Formation and Treatment Resistance. Oncogenesis 2022, 11, 46. [Google Scholar] [CrossRef]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Too Complex to Fail? Targeting Fatty Acid Metabolism for Cancer Therapy. Progress. Lipid Res. 2022, 85, 101143. [Google Scholar] [CrossRef]
- Zuzčák, M.; Trnka, J. Cellular Metabolism in Pancreatic Cancer as a Tool for Prognosis and Treatment (Review). Int. J. Oncol. 2022, 61, 93. [Google Scholar] [CrossRef] [PubMed]
- Kiseleva, O.; Kurbatov, I.; Ilgisonis, E.; Poverennaya, E. Defining Blood Plasma and Serum Metabolome by GC-MS. Metabolites 2021, 12, 15. [Google Scholar] [CrossRef]
- Kromrey, M.-L.; Bülow, R.; Hübner, J.; Paperlein, C.; Lerch, M.M.; Ittermann, T.; Völzke, H.; Mayerle, J.; Kühn, J.-P. Prospective Study on the Incidence, Prevalence and 5-Year Pancreatic-Related Mortality of Pancreatic Cysts in a Population-Based Study. Gut 2018, 67, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Zambirinis, C.P.; Pushalkar, S.; Saxena, D.; Miller, G. Pancreatic Cancer, Inflammation, and Microbiome. Cancer J. 2014, 20, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; DiVittore, N.A.; Young, M.M.; Jia, Z.; Xie, K.; Ritty, T.M.; Kester, M.; Fox, T.E. Altered Sphingolipid Metabolism in Patients with Metastatic Pancreatic Cancer. Biomolecules 2013, 3, 435–448. [Google Scholar] [CrossRef] [Green Version]
- Morad, S.A.F.; Cabot, M.C. Pancreatic Cancer and Sphingolipids. In Bioactive Sphingolipids in Cancer Biology and Therapy; Hannun, Y.A., Luberto, C., Mao, C., Obeid, L.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 211–233. ISBN 978-3-319-20750-6. [Google Scholar]
- Xu, R.; Yang, J.; Ren, B.; Wang, H.; Yang, G.; Chen, Y.; You, L.; Zhao, Y. Reprogramming of Amino Acid Metabolism in Pancreatic Cancer: Recent Advances and Therapeutic Strategies. Front. Oncol. 2020, 10, 572722. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [Green Version]
- Nakkina, S.P.; Gitto, S.B.; Pandey, V.; Parikh, J.G.; Geerts, D.; Maurer, H.C.; Olive, K.P.; Phanstiel, O.; Altomare, D.A. Differential Expression of Polyamine Pathways in Human Pancreatic Tumor Progression and Effects of Polyamine Blockade on Tumor Microenvironment. Cancers 2021, 13, 6391. [Google Scholar] [CrossRef]
- Novita Sari, I.; Setiawan, T.; Seock Kim, K.; Toni Wijaya, Y.; Won Cho, K.; Young Kwon, H. Metabolism and Function of Polyamines in Cancer Progression. Cancer Lett. 2021, 519, 91–104. [Google Scholar] [CrossRef]
- Sharma, A.; Chari, S.T. Pancreatic Cancer and Diabetes Mellitus. Curr. Treat. Options Gastroenterol. 2018, 16, 466–478. [Google Scholar] [CrossRef]
- Gouirand, V.; Gicquel, T.; Lien, E.C.; Jaune-Pons, E.; Da Costa, Q.; Finetti, P.; Metay, E.; Duluc, C.; Mayers, J.R.; Audebert, S.; et al. Ketogenic HMG-CoA Lyase and Its Product β-Hydroxybutyrate Promote Pancreatic Cancer Progression. EMBO J. 2022, 41, e110466. [Google Scholar] [CrossRef] [PubMed]
- Herner, A.; Sauliunaite, D.; Michalski, C.W.; Erkan, M.; De Oliveira, T.; Abiatari, I.; Kong, B.; Esposito, I.; Friess, H.; Kleeff, J. Glutamate Increases Pancreatic Cancer Cell Invasion and Migration via AMPA Receptor Activation and Kras-MAPK Signaling. Int. J. Cancer 2011, 129, 2349–2359. [Google Scholar] [CrossRef]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New Aspects of Amino Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Yang, J.-S.; Wang, C.-C.; Qiu, J.-D.; Ren, B.; You, L. Arginine Metabolism: A Potential Target in Pancreatic Cancer Therapy. Chin. Med. J. 2021, 134, 28–37. [Google Scholar] [CrossRef]
- Phanstiel IV, O. An Overview of Polyamine Metabolism in Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2018, 142, 1968–1976. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; He, P.; Gaida, M.; Yang, S.; Schetter, A.J.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Yfantis, H.; Lee, D.; et al. Inducible Nitric Oxide Synthase Enhances Disease Aggressiveness in Pancreatic Cancer. Oncotarget 2016, 7, 52993–53004. [Google Scholar] [CrossRef] [Green Version]
- Apiz Saab, J.J.; Dzierozynski, L.N.; Jonker, P.B.; AminiTabrizi, R.; Shah, H.; Menjivar, R.E.; Scott, A.J.; Nwosu, Z.C.; Zhu, Z.; Chen, R.N.; et al. Pancreatic Tumors Exhibit Myeloid-Driven Amino Acid Stress and Upregulate Arginine Biosynthesis. eLife 2023, 12, e81289. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.-F.; Chow, H.; Choi, S.; Leung, Y.-C. Arginine Deprivation Inhibits Pancreatic Cancer Cell Migration, Invasion and EMT via the down Regulation of Snail, Slug, Twist, and MMP1/9. J. Physiol. Biochem. 2020, 76, 73–83. [Google Scholar] [CrossRef]
- Zou, S.; Wang, X.; Liu, P.; Ke, C.; Xu, S. Arginine Metabolism and Deprivation in Cancer Therapy. Biomed. Pharmacother. 2019, 118, 109210. [Google Scholar] [CrossRef]
- Zaytouni, T.; Tsai, P.-Y.; Hitchcock, D.S.; DuBois, C.D.; Freinkman, E.; Lin, L.; Morales-Oyarvide, V.; Lenehan, P.J.; Wolpin, B.M.; Mino-Kenudson, M.; et al. Critical Role for Arginase 2 in Obesity-Associated Pancreatic Cancer. Nat. Commun. 2017, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Olivares, O.; Mayers, J.R.; Gouirand, V.; Torrence, M.E.; Gicquel, T.; Borge, L.; Lac, S.; Roques, J.; Lavaut, M.-N.; Berthezène, P.; et al. Collagen-Derived Proline Promotes Pancreatic Ductal Adenocarcinoma Cell Survival under Nutrient Limited Conditions. Nat. Commun. 2017, 8, 16031. [Google Scholar] [CrossRef]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Erratum: Pancreatic Stellate Cells Support Tumour Metabolism through Autophagic Alanine Secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Parker, S.J.; Amendola, C.R.; Hollinshead, K.E.R.; Yu, Q.; Yamamoto, K.; Encarnación-Rosado, J.; Rose, R.E.; LaRue, M.M.; Sohn, A.S.W.; Biancur, D.E.; et al. Selective Alanine Transporter Utilization Creates a Targetable Metabolic Niche in Pancreatic Cancer. Cancer Discov. 2020, 10, 1018–1037. [Google Scholar] [CrossRef]
- Li, F.; He, C.; Yao, H.; Zhao, Y.; Ye, X.; Zhou, S.; Zou, J.; Li, Y.; Li, J.; Chen, S.; et al. Glutamate from Nerve Cells Promotes Perineural Invasion in Pancreatic Cancer by Regulating Tumor Glycolysis through HK2 MRNA-M6A Modification. Pharmacol. Res. 2023, 187, 106555. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Hui, S.; Ghergurovich, J.M.; Fan, J.; Intlekofer, A.M.; White, R.M.; Rabinowitz, J.D.; Thompson, C.B.; Zhang, J. As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid. Cell Metab. 2018, 27, 428–438.e5. [Google Scholar] [CrossRef] [Green Version]
- Raho, S.; Capobianco, L.; Malivindi, R.; Vozza, A.; Piazzolla, C.; De Leonardis, F.; Gorgoglione, R.; Scarcia, P.; Pezzuto, F.; Agrimi, G.; et al. KRAS-Regulated Glutamine Metabolism Requires UCP2-Mediated Aspartate Transport to Support Pancreatic Cancer Growth. Nat. Metab. 2020, 2, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Anglin, J.; Zavareh, R.B.; Sander, P.N.; Haldar, D.; Mullarky, E.; Cantley, L.C.; Kimmelman, A.C.; Lyssiotis, C.A.; Lairson, L.L. Discovery and Optimization of Aspartate Aminotransferase 1 Inhibitors to Target Redox Balance in Pancreatic Ductal Adenocarcinoma. Bioorg. Med. Chem. Lett. 2018, 28, 2675–2678. [Google Scholar] [CrossRef]
- Geeraerts, S.L.; Heylen, E.; De Keersmaecker, K.; Kampen, K.R. The Ins and Outs of Serine and Glycine Metabolism in Cancer. Nat. Metab. 2021, 3, 131–141. [Google Scholar] [CrossRef] [PubMed]
- de Koning, T.J.; Snell, K.; Duran, M.; Berger, R.; Poll-The, B.-T.; Surtees, R. L-Serine in Disease and Development. Biochem. J. 2003, 371, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yu, H.; Ge, M.; Zhan, Q.; Huang, R.; Ji, X.; Liang, X.; Zhou, X. Upregulation of Phosphoserine Phosphatase Contributes to Tumor Progression and Predicts Poor Prognosis in Non-Small Cell Lung Cancer Patients. Thorac. Cancer 2019, 10, 1203–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and Glycine Metabolism in Cancer. Trends Biochem. Sciences 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Teoh, S.T.; Ensink, E.; Ogrodzinski, M.P.; Yang, C.; Vazquez, A.I.; Lunt, S.Y. Cysteine Catabolism and the Serine Biosynthesis Pathway Support Pyruvate Production during Pyruvate Kinase Knockdown in Pancreatic Cancer Cells. Cancer Metab. 2019, 7, 13. [Google Scholar] [CrossRef] [Green Version]
- Contorno, S.; Darienzo, R.E.; Tannenbaum, R. Evaluation of Aromatic Amino Acids as Potential Biomarkers in Breast Cancer by Raman Spectroscopy Analysis. Sci. Rep. 2021, 11, 1698. [Google Scholar] [CrossRef]
- Sikalidis, A.K. Amino Acids and Immune Response: A Role for Cysteine, Glutamine, Phenylalanine, Tryptophan and Arginine in T-Cell Function and Cancer? Pathol. Oncol. Res. 2015, 21, 9–17. [Google Scholar] [CrossRef]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of Mass Spectrometry–Based Metabolomics in Cancer Research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef] [Green Version]
- Peyraud, F.; Guegan, J.-P.; Bodet, D.; Cousin, S.; Bessede, A.; Italiano, A. Targeting Tryptophan Catabolism in Cancer Immunotherapy Era: Challenges and Perspectives. Front. Immunol. 2022, 13, 807271. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, Q.-Q.; Liu, Y.-M.; Yang, B. T Cells in Pancreatic Cancer Stroma: Tryptophan Metabolism Plays an Important Role in Immunoregulation. World J. Gastroenterol. 2023, 29, 2701–2703. [Google Scholar] [CrossRef]
- Deng, S.; Yang, X.; Lassus, H.; Liang, S.; Kaur, S.; Ye, Q.; Li, C.; Wang, L.-P.; Roby, K.F.; Orsulic, S.; et al. Distinct Expression Levels and Patterns of Stem Cell Marker, Aldehyde Dehydrogenase Isoform 1 (ALDH1), in Human Epithelial Cancers. PLoS ONE 2010, 5, e10277. [Google Scholar] [CrossRef]
- Lai, H.-S.; Lee, J.-C.; Lee, P.-H.; Wang, S.-T.; Chen, W.-J. Plasma Free Amino Acid Profile in Cancer Patients. Semin. Cancer Biol. 2005, 15, 267–276. [Google Scholar] [CrossRef]
- Yamamoto, K.; Iwadate, D.; Kato, H.; Nakai, Y.; Tateishi, K.; Fujishiro, M. Targeting the Metabolic Rewiring in Pancreatic Cancer and Its Tumor Microenvironment. Cancers 2022, 14, 4351. [Google Scholar] [CrossRef]
- Olson, K.A.; Schell, J.C.; Rutter, J. Pyruvate and Metabolic Flexibility: Illuminating a Path Toward Selective Cancer Therapies. Trends Biochem. Sci. 2016, 41, 219–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9, 1143. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.A.; Al-Sakkaf, K.; Shait Mohammed, M.R.; Dallol, A.; Al-Maghrabi, J.; Aldahlawi, A.; Ashoor, S.; Maamra, M.; Ragoussis, J.; Wu, W.; et al. Integration of Transcriptome and Metabolome Provides Unique Insights to Pathways Associated With Obese Breast Cancer Patients. Front. Oncol. 2020, 10, 804. [Google Scholar] [CrossRef]
- Muranaka, H.; Hendifar, A.; Osipov, A.; Moshayedi, N.; Placencio-Hickok, V.; Tatonetti, N.; Stotland, A.; Parker, S.; Van Eyk, J.; Pandol, S.J.; et al. Plasma Metabolomics Predicts Chemotherapy Response in Advanced Pancreatic Cancer. Cancers 2023, 15, 3020. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Hui, R.; Nouws, J.; Sauler, M.; Zeng, T.; Wu, Q. Untargeted Metabolomics Analysis of Esophageal Squamous Cell Cancer Progression. J. Transl. Med. 2022, 20, 127. [Google Scholar] [CrossRef]
- Kaoutari, A.E.; Fraunhoffer, N.A.; Hoare, O.; Teyssedou, C.; Soubeyran, P.; Gayet, O.; Roques, J.; Lomberk, G.; Urrutia, R.; Dusetti, N.; et al. Metabolomic Profiling of Pancreatic Adenocarcinoma Reveals Key Features Driving Clinical Outcome and Drug Resistance. eBioMedicine 2021, 66, 103332. [Google Scholar] [CrossRef]
- Wang, G.; Yao, H.; Gong, Y.; Lu, Z.; Pang, R.; Li, Y.; Yuan, Y.; Song, H.; Liu, J.; Jin, Y.; et al. Metabolic Detection and Systems Analyses of Pancreatic Ductal Adenocarcinoma through Machine Learning, Lipidomics, and Multi-Omics. Sci. Adv. 2021, 7, eabh2724. [Google Scholar] [CrossRef]
- Mahajan, U.M.; Alnatsha, A.; Li, Q.; Oehrle, B.; Weiss, F.-U.; Sendler, M.; Distler, M.; Uhl, W.; Fahlbusch, T.; Goni, E.; et al. Plasma Metabolome Profiling Identifies Metabolic Subtypes of Pancreatic Ductal Adenocarcinoma. Cells 2021, 10, 1821. [Google Scholar] [CrossRef]
- Giussani, P.; Tringali, C.; Riboni, L.; Viani, P.; Venerando, B. Sphingolipids: Key Regulators of Apoptosis and Pivotal Players in Cancer Drug Resistance. Int. J. Mol. Sci. 2014, 15, 4356–4392. [Google Scholar] [CrossRef] [Green Version]



| Study | Cohort Country | Samples | Data Set | Study Groups | Cancer Stage (n) | Age Mean | M/F |
|---|---|---|---|---|---|---|---|
| [12] | China | Tissue | Discovery | PDAC (51) | I (13), II (19), III (12), IV (7) | 60.9 | 29/22 |
| Precursor lesions (14): IPMN (3), MCN (4), SCN (7) | - | 55.35 | 8/6 | ||||
| Paired nontumor pancreatic tissue (40) | - | 63.35 | 23/17 | ||||
| Serum | Training | PDAC (80) | I (17), II (19), III (23), IV (21) | 61.2 | 48/32 | ||
| Precursor lesions (36): IPMN (15), MCN (9), SCN (12) | - | 58.05 | 23/13 | ||||
| Healthy subjects (48) | - | 60.35 | 31/17 | ||||
| Serum | Validation | PDAC (22) | I (8), II (14) | 66.91 | 11/11 | ||
| Precursor lesions (27): IPMN (6), MCN (7), SCN (14) | - | 59.15 | 15/12 | ||||
| Healthy subjects (27) | - | 58.37 | 17/10 | ||||
| [23] | China | Tissue | Discovery | PDAC (15) | - | NR | NR |
| Benign pancreatic disease (13): Cystadenoma and congenital cyst | NR | NR | |||||
| [24] | United States | Tissue and plasma | Training (tissue), validation (plasma) | PDAC (19) | IA, IB, IIA | 61.57 | 13/6 |
| Benign pancreatic disease (15): pancreatitis and pancreatic cystic neoplasms | 50.73 | 6/9 | |||||
| Precursor lesions (20): IPMN (14), MCN (3), others (3) | - | 57.9 | 7/13 | ||||
| Colorectal adenocarcinoma (28) | - | 60 | NR | ||||
| [25] | China | Serum | Whole data | Unresectable PDAC (36) | III (9), IV (27) | 63 | NR |
| Resectable PDAC (36) | I(30), II (6) | 59 | NR | ||||
| [26] | Czech Republic | Serum | Training (Phase III) | PDAC (430) | T1, T2, T3, T4 | NR | 219/211 |
| Healthy subjects (246) | - | NR | 122/124 | ||||
| Pancreatitis (22) | - | NR | 13/9 | ||||
| Validation (Phase III) | PDAC (116) | T1, T2, T3, T4 | NR | 56/60 | |||
| Healthy subjects (16) | - | NR | 6/10 | ||||
| [27] | Czech Republic | Plasma | Whole data | PDAC (43) | - | 67 | 26/17 |
| Healthy subjects (29) | - | 63 | 10/19 | ||||
| T2DM (34) | - | 68 | 18/16 | ||||
| RODM (59) | - | 65 | 27/32 | ||||
| [28] | Sweden | Plasma | Whole data | PDAC (10) | - | 76 | 6/4 |
| IPMN LGD (20) | - | 68 | 10/10 | ||||
| IPMN HGD (10) | - | 74 | 3/7 | ||||
| SCNs (5) | - | 47 | 0/5 | ||||
| Cyst fluid | Whole data | PDAC (15) | - | 71 | 7/8 | ||
| IPMN LGD (29) | - | 70 | 12/17 | ||||
| IPMN HGD (8) | - | 76 | 3/5 | ||||
| SCNs (5) | - | 47 | 0/5 | ||||
| [29] | China | Plasma | Whole data | PDAC (26) | - | 64.74 | 14/12 |
| DM (27) | - | 56.93 | 14/13 | ||||
| Healthy subjects (23) | - | 60.22 | 12/11 | ||||
| [30] | United States | Plasma | Training (Cohort 1) | PDAC (20) | IB (2); IIA (1); IIB (7); IV (10) | 70.4 | 10/10 |
| Healthy subjects (10) | - | 60.2 | 4/6 | ||||
| Healthy subjects (60) | - | 62.0 | 30/30 | ||||
| Chronic pancreatitis (10) | - | 61.6 | 6/4 | ||||
| Training (Cohort 2) | PDAC (9) | IA (2); IIA (2); IIB (2); IV (4) | 73.1 | 3/6 | |||
| IPMNs (34), MNC (11), SCN (6) | - | 62.2 | 19/32 | ||||
| Validation | PDAC (39) | IA (6); IB (10); resectable No TNM data (22) | 62.0 | 21/18 | |||
| Healthy subjects (82) | - | 62.8 | 43/39 | ||||
| [31] | United States | Plasma | Whole data | IPMN (10) | - | 66 | 6/4 |
| Localized PDAC (10) | - | 63 | 4/6 | ||||
| Locally advanced PDAC with nodal disease (10) | - | 70 | 6/4 | ||||
| Unresectable metastatic (10) | - | 64 | 6/4 | ||||
| PNET (10) | - | 64 | 8/2 | ||||
| [32] | Lithuania | Plasma | Whole data | PDAC (50) | IA (4); IB (4); IIA (5); IIB (21); III (16) | 65.5 | 28/22 |
| Other pancreatic cancers (18) | - | 63.5 | 10/8 | ||||
| Chronic pancreatitis (7) | - | 58 | 4/3 | ||||
| [33] | Germany | Plasma | Exploratory study | PDAC (34) | IB (1); IIA (4); IIB (8); III (11) and IV (10) | 64 | 15/19 |
| Chronic pancreatitis (43) | - | 50 | 36/7 | ||||
| Liver cirrhosis (20) | - | 56 | 15/5 | ||||
| Healthy subjects and non-pancreatic disease control blood donors (104) | - | 53 | 49/55 | ||||
| Serum and plasma | Training | PDAC (158): serum (80) and plasma (78) | IA (2); IB (3); IIA (18); IIB (59); III (22); IV (54) | 70 | 102/56 | ||
| Chronic pancreatitis (159): serum (79) and plasma (80) | - | 50 | 136/23 | ||||
| Liver cirrhosis (80): serum (80) | - | 61 | 60/20 | ||||
| Non-pancreatic disease control blood donors (77): serum (77) | - | 55 | 51/26 | ||||
| Plasma | Validation | PDAC (79) | IA (1); IIA (11); IIB (28); III (26); IV (13) | 69 | 37/42 | ||
| Chronic pancreatitis (80) | - | 51 | 62/18 | ||||
| Non-pancreatic disease preoperative patients (80) | - | 68 | 42/38 | ||||
| [34] | Japan | Urine | Validation | PDAC (67) | - | 72 | 48/19 |
| Pancreatitis (11) | - | 67 | 8/2 | ||||
| Healthy subjects (9) | - | 69 | 9/0 | ||||
| [35] | Japan | Saliva | Development | PDAC (39) | III (6), IVa (12), IVb (21) | 66.1 | 21/18 |
| Chronic pancreatitis (14) | - | 51.1 | 11/3 | ||||
| Healthy subjects (26) | - | 50.8 | 13/13 |
| Study | Sample | Metabolomics Approach | Data Set | More Relevant Comparison | Results (Metabolites) | AUC (CI-CI) | Sens. (%) | Spec. (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|
| [23] | Tissue | HR-MAS NMR | Discovery | PDAC vs. benign pancreatic disease | Lactate and ethanol > in PDAC. Methylene of lipid (L-CH2), myo-inositol, phosphocholine and glycerophosphocholine < in PDAC | NR | NR | NR | NR |
| [24] | Tissue | UHPLC/ESI-Q-TOF-MS | Discovery | Early PDAC vs. benign pancreatic disease | 6-metabolite panel: 5-hydroxytryptophan, LysoPE (18:2), PC (16:0/16:0), PC (18:0/22:4), PE (17:0) and SM (d18:1/16:0) | 0.95 (0.78–1) | 90 | 85 | NR |
| High-risk PDAC vs. benign pancreatic disease | 6-metabolite panel: 5-hydroxytryptophan, LysoPE (18:2), PC (16:0/16:0), PC (18:0/22:4), PE (17:0) and SM (d18:1/16:0) | 0.46 (0.21–0.73) | NR | NR | NR | ||||
| 12 metabolites: 1-Indanol (with 12 different m/z) | 0.836 (0.57–0.98) | NR | NR | NR | |||||
| Plasma | MRM-MS | Validation | Early PDAC vs. benign pancreatic disease | 6-metabolite panel: 5-hydroxytryptophan, LysoPE (18:2), PC (16:0/16:0), PC (18:0/22:4), PE (17:0) and SM (d18:1/16:0) | Failure in the analysis due to discordant metabolite abundance results with respect to tissue. | NR | NR | NR | |
| [27] | Plasma | 1H NMR | Whole data | PDAC vs. healthy subjects | Increased: 3-hydroxybutyrate and mannose Decreased: creatine, ornithine, alanine, uridine, serine, histidine, carnitine, glutamine, glycine, threonine, lysine and methionine | NR | NR | NR | NR |
| T2DM vs. healthy subjects | Increased: glucose Decreased: 18 metabolites (ornithine, uridine, histidine and glutamine) | NR | NR | NR | NR | ||||
| PDAC vs. T2DM | Increased: 3-hydroxybutyrate, propylene glycol, mannose, propionate, glutamate and tryptophan Decreased: creatine, alanine, valine, proline and lysine | NR | NR | NR | NR | ||||
| PDAC vs. (T2DM and healthy subjects) | Increased: 3-hydroxybutyrate, mannose and glutamate Decreased: creatine, alanine, valine, proline and lysine | NR | NR | NR | NR | ||||
| [28] | Plasma | UHPLC/MS | Whole data | (HGD or PDAC) vs. (LGD or SCNs) | Bacterial metabolite trimethylamine oxide (9.12 µM) | 0.82 (0.65–0.94) | 80 | 90 | NR |
| Taurochenodeoxycholate (204 nM) | 0.73 (0.56–0.87) | 80 | 70 | NR | |||||
| [29] | Plasma | UHPLC/MS | Whole data | PDAC vs. (DM and healthy subjects) | Increased: lysoPC (22:6), lysoPC (20:3) and 1,2,4-nonadecanetriol Reduced: lysoPC (16:0) | NR | NR | NR | NR |
| (PDAC and DM) vs. healthy subjects | Increased: lysoPC (20:4), deoxyadenosine, asparaginyl-histidine and vaccenyl carnitine Reduced: phytal, 2 (R)-hydroxydocosanoic acid, behenic acid, catelaidic acid, 2-hydroxyphytanic acid, phytosphingosine, cerebronic acid, docosanamide and eicosenoic acid | NR | NR | NR | NR | ||||
| PDAC vs. healthy subjects | Combination 1: lysoPC (22:6), catelaidic acid, cerebronic acid, docosanamide and asparaginyl-Histidine | 0.882 (0.846–0.918) | 81.6 (76.0–87.2) | 87.3 (83.4–91.2) | 85.2 (82.1–88.3) | ||||
| Combination 2: lysoPC (16:0), catelaidic acid, cerebronic acid, nonadecanetriol and asparaginyl-histidine | 0.974 (0.958–0.991) | 89.0 (84.7–93.3) | 90.6 (86.1–95.1) | 88.6 (86.4–90.9) | |||||
| Combination 3: lysoPC (22:6), catelaidic acid, cerebronic acid, docosanamide and asparaginyl-histidine | 0.879 (0.848–0.909) | 83.4 (78.1–88.7) | 89.6 (86.0–93.2) | 86.5 (84.3–88.8) | |||||
| Combination 4: lysoPC (16:0), lysoPC (16:1), lysoPC (22:6) and lysoPC (20:3) | 0.860 (0.823–0.896) | 84.8 (78.5–91.1) | 83.1 (77.8–88.3) | 83.7 (80.1–87.4) | |||||
| Combination 5: lysoPC (16:0), lysoPC (16:1), lysoPC (22:6) and lysoPC (20:3), catelaidic acid, cerebronic acid, docosanamide, nonadecanetriol and asparaginyl-histidine | 0.919 (0.887–0.952) | 89.4 (85.3–93.5) | 77.3 (71.4–83.2) | 84.1 (80.8–87.4) | |||||
| CA19-9 | 0.821 (0.765–0.874) | 79.1 (74.5–82.6) | 82.6 (76.5–89.4) | 81.3 (77.8–83.4) | |||||
| PDAC vs. DM | Combination 1 (same as PDAC vs. healthy subjects) | 0.586 (0.534–0.638) | 50.3 (43.9–56.6) | 62.9 (57.5–68.3) | 58.6 (55.6–61.5) | ||||
| Combination 2 (same as PDAC vs. healthy subjects) | 0.631 (0.580–0.682) | 54.7 (49.3–60.0) | 61.3 (54.4–68.3) | 54.7 (52.2–57.2) | |||||
| Combination 3 (same as PDAC vs. healthy subjects) | 0.569 (0.516–0.622) | 46.3 (39.8–52.7) | 61.2 (54.3–68.1) | 54.7 (51.6–57.9) | |||||
| Combination 4 (same as PDAC vs. healthy subjects) | 0.723 (0.691–0.754) | 63.5 (58.9–68.1) | 69.6 (64.1–75.1) | 67.7 (64.1–71.3) | |||||
| Combination 5 (same as PDAC vs. healthy subjects) | 0.680 (0.632–0.729) | 58.9 (53.5–64.3) | 67.1 (60.9–73.2) | 65.9 (62.6–69.2) | |||||
| [30] | Plasma | UHPLC/MS | Training | PDAC vs. healthy subjects | 5-marker metabolite panel: (N1/N8)-acetylspermidine (AcSperm), DAS, lysoPC (18:0), lysoPC (20:3) and an indole-derivative | 0.903 (0.818–0.989) | NR | NR | NR |
| CA19-9 | 0.859 (0.743–0.975) | NR | NR | NR | |||||
| Protein (CA19-9, TIMP1 and LRG1) | 0.948 (0.883–1.000) | NR | NR | NR | |||||
| Protein (CA19-9, TIMP1 and LRG1) + metabolite multiplex panel | 0.972 (0.928–1.000) | NR | NR | NR | |||||
| Validation | PDAC vs. healthy subjects | Indole-derivative | 0.726 (0.631–0.822) | 23.1 (95% specificity) | 11.3 (95% sensitivity) | NR | |||
| LysoPC (18:0) | 0.842 (0.764–0.920) | 51.3 (95% specificity) | 26.3 (95% sensitivity) | NR | |||||
| LysoPC (20:3) | 0.841 (0.757–0.925) | 48.7 (95% specificity) | 11.3 (95% sensitivity) | NR | |||||
| AcSperm | 0.755 (0.659–0.852) | 33.3 (95% specificity) | 27.5 (95% sensitivity) | NR | |||||
| DAS | 0.801 (0.712–0.890) | 51.3 (95% specificity) | 27.5 (95% sensitivity) | NR | |||||
| 5-marker metabolite panel | 0.892 (0.828–0.956) | 66.7 (95% specificity) | 43.3 (95% sensitivity) | NR | |||||
| CA19-9 | 0.800 (0.708–0.891) | NR | NR | NR | |||||
| Protein (CA19-9, TIMP1 and LRG1) | 0.863 (0.782–0.946) | NR | NR | NR | |||||
| Protein (CA19-9, TIMP1 and LRG1) + metabolite multiplex panel | 0.924 (0.864–0.983) | NR | NR | NR | |||||
| [31] | Plasma | UHPLC/MS | Whole data | Higher correlation with disease state than CA19-9 | Four metabolites: lysine, propionyl-carnitine, C5-acylcarnitine and dodecanedioic acid | NR | NR | NR | NR |
| PNET | High: uric acid, methionine | NR | NR | NR | NR | ||||
| IPMN | High: amino acid | NR | NR | NR | NR | ||||
| Locally advanced PDAC | High: fatty acid and polyamines | NR | NR | NR | NR | ||||
| Metastatic PDAC | High: TCA cycle metabolites | NR | NR | NR | NR | ||||
| Local PDAC | No predominance of specific principal components | NR | NR | NR | NR | ||||
| [32] | Plasma | Ion-exchange chromatography | Whole data | PDAC vs. (OPC and chronic pancreatitis) | Ornithine, threonine, phenylalanine, glycine, arginine, histidine, glutamine, 3-methylhistidine and citruline | NR | NR | NR | NR |
| PDAC vs. OPC | Ornithine, threonine, phenylalanine, lysine, valine, arginine, histidine, asparagine, glutamine, 3-methylhistidine and citruline | NR | NR | NR | NR | ||||
| Different PDAC stages | Inverse correlation between plasma histidine concentrations and PDAC stage. U-shaped curves from stage I to stage IV were observed for tyrosine, proline, glycine, arginine, serine and threonine | NR | NR | NR | NR | ||||
| [33] | Plasma | GC-MS; LC–MS/MS; SPE-LC–MS/MS | Validation/test | PDAC (all stages) vs. chronic pancreatitis | Biomarker signature | 0.94 (0.91–0.97) | 89.9 (81–95.5) | 91.3 (82.8–96.4) | 90.6 (84.9–94.6) |
| CA19-9 | 0.85 | NR | NR | NR | |||||
| Resectable PDAC (stages IA-IIB) vs. chronic pancreatitis | Biomarker signature | 0.93 | 90.0 (76.3–97.2) | 91.3 (82.8–96.4) | 90.8 (84.2–95.3) | ||||
| CA19-9 | 0.84 | NR | NR | NR | |||||
| PDAC (all stages) vs. don-pancreatic controls | Biomarker signature | 0.9 | NR | NR | NR | ||||
| CA19-9 | 0.89 | NR | NR | NR | |||||
| Resectable PDAC (stages IA-IIB) vs. Non-pancreatic controls | Biomarker signature | 0.88 | NR | NR | NR | ||||
| CA19-9 | 0.88 | NR | NR | NR | |||||
| Plasma and serum | Training | PDAC (all stages) vs. chronic pancreatitis | Biomarker signature: CA19-9 and nine metabolites (Proline; SM (d18:2,C17:0); PC (C18:0,C22:6); isocitrate; sphinganine-1-phosphate (d18:0); histidine; pyruvate; Cer (d18:1,C24:0); SM (d17:1,C18:0)) | Plasma: 0.96 (0.93–0.98) Serum: 0.88 | Plasma: 94.9 (87–97) | Plasma: 85 | Plasma: 90 (86–91) | ||
| CA19-9 | Plasma: 0.88 Serum: 0.8 | NR | NR | NR | |||||
| Resectable PDAC (stages IA-IIB) vs. chronic pancreatitis | Biomarker signature | Plasma: 0.99 Serum: 0.81 | Plasma: 98.2 (93.3–99.4) | Plasma: 85 | Plasma: 91.6 (89.2–92.2) | ||||
| CA19-9 | Plasma: 0.91 Serum: 0.7 | NR | NR | NR | |||||
| PDAC (all stages) vs. liver cirrhosis | Biomarker signature | Serum: 0.87 | NR | NR | NR | ||||
| CA19-9 | Serum: 0.79 | NR | NR | NR | |||||
| Resectable PDAC (stages IA-IIB) vs. liver cirrhosis | Biomarker signature | Serum: 0.79 | NR | NR | NR | ||||
| CA19-9 | Serum: 0.7 | NR | NR | NR | |||||
| PDAC (all stages) vs. healthy subjects | Biomarker signature | Serum: 0.95 | NR | NR | NR | ||||
| CA19-9 | Serum: 0.88 | NR | NR | NR | |||||
| Resectable PDAC (stages IA-IIB) vs. healthy subjects | Biomarker signature | Serum: 0.87 | NR | NR | NR | ||||
| CA19-9 | Serum: 0.79 | NR | NR | NR | |||||
| [12] | Serum | UHPLC/Q-TOF-MS | Training | PDAC vs. healthy subjects | Proline, creatine and palmitic acid | 0.854 (0.842–0.865) | 80 | 79.2 | 79.7 |
| Proline, creatine and palmitic acid + 19-9 | 0.919 (0.911–0.928) | 82.5 | 89.6 | 85.2 | |||||
| Early PDAC vs. healthy subjects | Proline, creatine and palmitic acid | 0.880 (0.864–0.896) | 88.9 | 79.2 | 83.3 | ||||
| Proline, creatine and palmitic acid + CA19-9 | 0.900 (0.886–0.915) | 86.1 | 85.4 | 85.7 | |||||
| PDAC vs. precursor lesions | Proline, creatine and palmitic acid | 0.865 (0.800–0.931) | 76.3 | 86.1 | NR | ||||
| CA19-9 | 0.806 (0.719–0.892) | 75 | 86.1 | 72.2 | |||||
| Proline, creatine and palmitic acid + CA19-9 | 0.917 (0.868–0.966) | 86.3 | 86.1 | NR | |||||
| CA19-9 negative PDAC patients vs. healthy subjects | Proline, creatine and palmitic acid | 0.851 (0.840–0.863) | 75.4 | 70.1 | 72 | ||||
| Validation | Early PDAC vs. healthy subjects | Proline, creatine and palmitic acid | 0.83 (0.792–0.866) | 76.2 | 70.4 | 72.9 | |||
| Proline, creatine and palmitic acid + 19-9 | 0.949 (0.933–0.966) | 85.7 | 81.5 | 83.3 | |||||
| Early PDAC vs. precursor lesions | Proline, creatine and palmitic acid | 0.852 (0.736–0.967) | 86.4 | 77.8 | NR | ||||
| CA19-9 | 0.757 (0.616–0.897) | 77.3 | 74.1 | 73.5 | |||||
| Proline, creatine and palmitic acid + CA19-9 | 0.909 (0.825–0.993) | 81.8 | 88.9 | NR | |||||
| [25] | Serum | UHPLC/Q-TOF-MS | Whole data | Unresectable PDAC vs. resectable PDAC | Oleic acid | 0.965 (0.922–0.991) | NR | NR | NR |
| Linoleic acid | 0.979 (0.945–0.998) | ||||||||
| Palmitic acid | 0.984 (0.957–1) | ||||||||
| Linoelaidyl carnitine | 0.965 (0.918–0.992) | ||||||||
| 2-Octenedioic acid | 1 (1–1) | ||||||||
| 3R,7R-1,3,7-Octanetriol | 0.984 (0.949–1) | ||||||||
| LysoPE (P-16:0/0:0) | 0.981 (0.947–1) | ||||||||
| 3-Hydroxyanthranilic acid | 0.957 (0.902–0.989) | ||||||||
| [26] | Serum | UHPSFC/MS | Training and Validation | PDAC vs. healthy subjects | CA19-9 | 0.854 | 70.33 | 97.33 | 79.08 |
| Lipids | 0.983 | 95.97 | 90.46 | 94.18 | |||||
| CA19-9 + lipids | 0.989 | 95.97 | 92.75 | 94.93 | |||||
| Training | PDAC vs. healthy subjects | The lipid species with the highest relevance are SM (41:1), SM (42:1), Cer (41:1), Cer (42:1), SM (39:1), LysoPC (18:2) and PC (O-36:3) | NR | NR | NR | NR | |||
| [34] | Urine | HPLC | Validation | Normal pancreas, PDAC and pancreatitis | CA19-9 | NR | 74.6 | 83.3 | 76.5 |
| UP | NR | 55.2 | 70 | 58.6 | |||||
| CP | NR | 41.8 | 85 | 51.7 | |||||
| CA19-9 + UP | NR | 83.6 | 57.9 | 77.9 | |||||
| CA19-9 + CP | NR | 86.6 | 68.4 | 82.6 | |||||
| CA19-9 + UP + CP | NR | 89.6 | 52.6 | 81.4 | |||||
| [35] | Saliva | CE-TOF/MS | Development | Normal pancreas, PDAC and chronic pancreatitis | Alanine, N1-acetylspermidine, 2-oxobutyrate and 2-hydroxybutyrate | 0.887 (0.784–0.944) | NR | NR | NR |
| [28] | Cyst fluid | UHPLC/MS | Whole data | (SCN and LGD) vs. (HGD and PDAC) | Acyl-C4 (0.237 µM) | 0.83 (0.69–0.93) | 80 | 80 | NR |
| Acyl-C4-OH (0.0751 µM) | 0.79 (0.67–0.90) | 80 | 80 | NR | |||||
| Acyl-C2 (6.94 µM) | 0.78 (0.65–0.89) | 80 | 70 | NR | |||||
| Acyl-C6 (0.0374 µM) | 0.77 (0.63–0.89) | 80 | 80 | NR | |||||
| Choline (7.06 µM) | 0.78 (0.65–0.89) | 70 | 80 | NR | |||||
| Succinate (2.89 µM) | 0.80 (0.67–0.91) | 90 | 70 | NR | |||||
| Fumarate (1.11 µM) | 0.76 (0.62–0.89) | 70 | 80 | NR | |||||
| Malate (20.5 µM) | 0.78 (0.64–0.90) | 70 | 80 | NR | |||||
| SCN vs. PDAC | 5-Oxoproline (247 µM) | 1 (1–1) | NR | ||||||
| Glutamine (264 µM) | 1 (0.935–1) | NR | |||||||
| Ethanolamine phosphate | 1 (0.935–1) | 100 | 90 | NR | |||||
| D-Glucose | 0.971 (0.871–1) | 100 | 90 | NR | |||||
| HGD vs. PDAC | Indole | 0.89 (0.655–1) | 90 | 90 | NR | ||||
| L-Adrenaline | 0.85 (0.63–0.98) | 90 | 70 | NR | |||||
| Malate | 0.78 (0.52–0.96) | 70 | 80 | NR | |||||
| S-Adenosyl-L-methionine | 0.83 (0.61–0.96) | 70 | 90 | NR | |||||
| Dopamine | 0.83 (0.635–0.96) | 60 | 90 | NR | |||||
| Tryptophan | 0.82 (0.61–0.96) | 80 | 70 | NR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perazzoli, G.; García-Valdeavero, O.M.; Peña, M.; Prados, J.; Melguizo, C.; Jiménez-Luna, C. Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review. Metabolites 2023, 13, 872. https://doi.org/10.3390/metabo13070872
Perazzoli G, García-Valdeavero OM, Peña M, Prados J, Melguizo C, Jiménez-Luna C. Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review. Metabolites. 2023; 13(7):872. https://doi.org/10.3390/metabo13070872
Chicago/Turabian StylePerazzoli, Gloria, Olga M. García-Valdeavero, Mercedes Peña, Jose Prados, Consolación Melguizo, and Cristina Jiménez-Luna. 2023. "Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review" Metabolites 13, no. 7: 872. https://doi.org/10.3390/metabo13070872
APA StylePerazzoli, G., García-Valdeavero, O. M., Peña, M., Prados, J., Melguizo, C., & Jiménez-Luna, C. (2023). Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review. Metabolites, 13(7), 872. https://doi.org/10.3390/metabo13070872