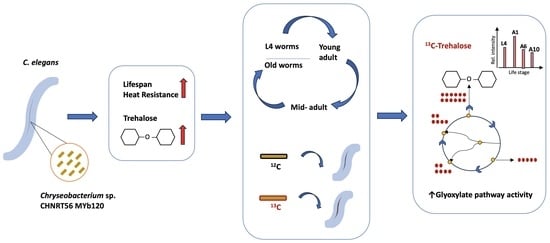
The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Growth Culture
2.2. Maintenance of C. elegans Worms
2.3. Lifespan Assay
2.4. Worm Motility (Head Thrashing) Assay
2.5. Worm Heat Resistance Assay
2.6. Global Metabolomics and Stable Isotope Labeling of C. elegans Worms
2.7. Metabolite Extraction
2.8. LC-MS and MS/MS
2.9. Statistical Analysis
2.10. Functional Analysis
3. Results
3.1. Chryseobacterium sp. CHNTR56 MYb120 Increases Lifespan and Heat-Stress Resistance but Reduces Motility Functions in C. elegans
3.2. The Influence of Native Member Chryseobacterium sp. CHNTR56 MYb120 on the Metabolome of the Young Adult C. elegans Worm
3.3. Chryseobacterium sp. CHNTR56 MYb120 Upregulates the Abundance of Central Carbon Metabolism Intermediates in C. elegans during the Early Life Stages
3.4. Isotope Labeling Analysis Reveals the Influence of Chryseobacterium sp. CHNTR56 MYb120 on the Upregulation of Desiccation-Tolerance Pathways in Worms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adav, S.S.; Wang, Y. Metabolomics signatures of aging: Recent advances. Aging Dis. 2021, 12, 646. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Quipildor, G.F.; Tabrizian, T.; Novaj, A.; Guan, F.; Walters, R.O.; Delahaye, F.; Hubbard, G.B.; Ikeno, Y.; Ejima, K.; et al. Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat. Commun. 2018, 9, 2394. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Shah, P.P.; Nativio, R.; Berger, S.L. Epigenetic mechanisms of longevity and aging. Cell 2016, 166, 822–839. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Ramanathan, A. The Aging Metabolome—Biomarkers to Hub Metabolites. Proteomics 2020, 20, 1800407. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Shanahan, F.; O’Toole, P.W. The gut microbiome as a modulator of healthy ageing. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 565–584. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Wang, R.; Yin, Y.; Li, J.; Wang, H.; Lv, W.; Gao, Y.; Wang, T.; Zhong, Y.; Zhou, Z.; Cai, Y.; et al. Global stable-isotope tracing metabolomics reveals system-wide metabolic alternations in aging Drosophila. Nat. Commun. 2022, 13, 3518. [Google Scholar] [CrossRef]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef]
- Dorman, J.B.; Albinder, B.; Shroyer, T.; Kenyon, C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 1995, 141, 1399–1406. [Google Scholar] [CrossRef]
- Dall, K.B.; Færgeman, N.J. Metabolic regulation of lifespan from a C. elegans perspective. Genes Nutr. 2019, 14, 25. [Google Scholar] [CrossRef]
- Fuchs, S.; Bundy, J.G.; Davies, S.K.; Viney, J.M.; Swire, J.S.; Leroi, A.M. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.J.; Zarse, K.; Voigt, A.; Urban, N.; Birringer, M.; Ristow, M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007, 6, 280–293. [Google Scholar] [CrossRef]
- Lamitina, T.; Kitaoka, S.; Morielli, A.D.; Zhao, F.-Q. FGT-1 Is a Mammalian GLUT2-Like Facilitative Glucose Transporter in Caenorhabditis elegans Whose Malfunction Induces Fat Accumulation in Intestinal Cells. PLoS ONE 2013, 8, e68475. [Google Scholar] [CrossRef]
- Seo, Y.; Kingsley, S.; Walker, G.; Mondoux, M.A.; Tissenbaum, H.A. Metabolic shift from glycogen to trehalose promotes lifespan and healthspan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2018, 115, E2791–E2800. [Google Scholar] [CrossRef]
- Tauffenberger, A.; Vaccaro, A.; Parker, J.A. Fragile lifespan expansion by dietary mitohormesis in C. elegans. Aging 2016, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Ristow, M. Lipid and Carbohydrate Metabolism in Caenorhabditis elegans. Genetics 2017, 207, 413–446. [Google Scholar] [CrossRef] [PubMed]
- Noronha, S.; Yeh, H.; Spande, T.; Shiloach, J. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using 13C-NMR/MS. Biotechnol. Bioeng. 2000, 68, 316–327. [Google Scholar] [CrossRef]
- Erkut, C.; Gade, V.R.; Laxman, S.; Kurzchalia, T.V. The glyoxylate shunt is essential for desiccation tolerance in C. elegans and budding yeast. eLife 2016, 5, e13614. [Google Scholar] [CrossRef]
- Cabreiro, F.; Au, C.; Leung, K.-Y.; Vergara-Irigaray, N.; Cochemé, H.M.; Noori, T.; Weinkove, D.; Schuster, E.; Greene, N.D.; Gems, D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 2013, 153, 228–239. [Google Scholar] [CrossRef]
- García-González, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell 2017, 169, 431–441.e438. [Google Scholar] [CrossRef]
- Dirksen, P.; Assié, A.; Zimmermann, J.; Zhang, F.; Tietje, A.M.; Marsh, S.A.; Félix, M.A.; Shapira, M.; Kaleta, C.; Schulenburg, H.; et al. CeMbio—The Caenorhabditis elegans Microbiome Resource. G3 Genes|Genomes|Genet. 2020, 10, 3025. [Google Scholar] [CrossRef] [PubMed]
- Dirksen, P.; Marsh, S.A.; Braker, I.; Heitland, N.; Wagner, S.; Nakad, R.; Mader, S.; Petersen, C.; Kowallik, V.; Rosenstiel, P.; et al. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Obeng, N.; Yang, W.; Pees, B.; Petersen, C.; Waschina, S.; Kissoyan, K.A.; Aidley, J.; Hoeppner, M.P.; Bunk, B. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J. 2020, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Petersen, C.; Pees, B.; Zimmermann, J.; Waschina, S.; Dirksen, P.; Rosenstiel, P.; Tholey, A.; Leippe, M.; Dierking, K. The inducible response of the nematode Caenorhabditis elegans to members of its natural microbiota across development and adult life. Front. Microbiol. 2019, 10, 1793. [Google Scholar] [CrossRef]
- Haçariz, O.; Viau, C.; Karimian, F.; Xia, J. The symbiotic relationship between Caenorhabditis elegans and members of its microbiome contributes to worm fitness and lifespan extension. BMC Genom. 2021, 22, 364. [Google Scholar] [CrossRef]
- Haçariz, O.; Viau, C.; Gu, X.; Xia, J. Native Microbiome Members of C. elegans Act Synergistically in Biosynthesis of Pyridoxal 5′-Phosphate. Metabolites 2022, 12, 172. [Google Scholar] [CrossRef]
- Kerr, R.A.; Roux, A.E.; Goudeau, J.F.; Kenyon, C. The C. elegans Observatory: High-throughput exploration of behavioral aging. Front. Aging 2022, 3, 932656. [Google Scholar] [CrossRef]
- Gems, D.; Riddle, D.L. Defining wild-type life span in Caenorhabditis elegans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2000, 55, B215–B219. [Google Scholar] [CrossRef]
- Honda, Y.; Tanaka, M.; Honda, S. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell 2010, 9, 558–569. [Google Scholar] [CrossRef]
- Han, S.K.; Lee, D.; Lee, H.; Kim, D.; Son, H.G.; Yang, J.-S.; Lee, S.-J.V.; Kim, S. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 2016, 7, 56147. [Google Scholar] [CrossRef]
- Perez, C.L.; Van Gilst, M.R. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008, 8, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Barardo, D.; Thornton, D.; Thoppil, H.; Walsh, M.; Sharifi, S.; Ferreira, S.; Anžič, A.; Fernandes, M.; Monteiro, P.; Grum, T. The DrugAge database of aging-related drugs. Aging Cell 2017, 16, 594–597. [Google Scholar] [CrossRef]
- Llufrio, E.M.; Cho, K.; Patti, G.J. Systems-level analysis of isotopic labeling in untargeted metabolomic data by X13CMS. Nat. Protoc. 2019, 14, 1970–1990. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Davies, S.K.; Leroi, A.M.; Bundy, J.G. Fluorodeoxyuridine affects the identification of metabolic responses to daf-2 status in Caenorhabditis elegans. Mech. Ageing Dev. 2012, 133, 46–49. [Google Scholar] [CrossRef]
- Savion, N.; Levine, A.; Kotev-Emeth, S.; Bening Abu-Shach, U.; Broday, L. S-allylmercapto-N-acetylcysteine protects against oxidative stress and extends lifespan in Caenorhabditis elegans. PLoS ONE 2018, 13, e0194780. [Google Scholar] [CrossRef] [PubMed]
- Johnke, J.; Dirksen, P.; Schulenburg, H. Community assembly of the native C. elegans microbiome is influenced by time, substrate and individual bacterial taxa. Environ. Microbiol. 2020, 22, 1265–1279. [Google Scholar] [CrossRef]
- Virk, B.; Jia, J.; Maynard, C.A.; Raimundo, A.; Lefebvre, J.; Richards, S.A.; Chetina, N.; Liang, Y.; Helliwell, N.; Cipinska, M. Folate acts in E. coli to accelerate C. elegans aging independently of bacterial biosynthesis. Cell Rep. 2016, 14, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Virk, B.; Correia, G.; Dixon, D.P.; Feyst, I.; Jia, J.; Oberleitner, N.; Briggs, Z.; Hodge, E.; Edwards, R.; Ward, J. Excessive folate synthesis limits lifespan in the C. elegans: E. coliaging model. BMC Biol. 2012, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Holden, H.M.; Rayment, I.; Thoden, J.B. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 2003, 278, 43885–43888. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, C.; Wang, M.; Tran, P.; Mai, N.; Finley, J.W.; Heymsfield, S.B.; Greenway, F.L.; Li, Z.; Heber, D. Lower doses of fructose extend lifespan in Caenorhabditis elegans. J. Diet. Suppl. 2017, 14, 264–277. [Google Scholar] [CrossRef]
- Gao, A.W.; Smith, R.L.; van Weeghel, M.; Kamble, R.; Janssens, G.E.; Houtkooper, R.H. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 2018, 113, 128. [Google Scholar] [CrossRef]
- Copes, N.; Edwards, C.; Chaput, D.; Saifee, M.; Barjuca, I.; Nelson, D.; Paraggio, A.; Saad, P.; Lipps, D.; Stevens Jr, S.M. Metabolome and proteome changes with aging in Caenorhabditis elegans. Exp. Gerontol. 2015, 72, 67–84. [Google Scholar] [CrossRef]
- Edwards, C.B.; Copes, N.; Brito, A.G.; Canfield, J.; Bradshaw, P.C. Malate and fumarate extend lifespan in Caenorhabditis elegans. PLoS ONE 2013, 8, e58345. [Google Scholar] [CrossRef]
- Murima, P.; Zimmermann, M.; Chopra, T.; Pojer, F.; Fonti, G.; Dal Peraro, M.; Alonso, S.; Sauer, U.; Pethe, K.; McKinney, J.D. A rheostat mechanism governs the bifurcation of carbon flux in mycobacteria. Nat. Commun. 2016, 7, 12527. [Google Scholar] [CrossRef]
- Elbein, A.D.; Pan, Y.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef]
- Pellerone, F.; Archer, S.; Behm, C.; Grant, W.N.; Lacey, M.; Somerville, A. Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes. Int. J. Parasitol. 2003, 33, 1195–1206. [Google Scholar] [CrossRef]
- Hibshman, J.D.; Doan, A.E.; Moore, B.T.; Kaplan, R.E.; Hung, A.; Webster, A.K.; Bhatt, D.P.; Chitrakar, R.; Hirschey, M.D.; Baugh, L.R. daf-16/FoxO promotes gluconeogenesis and trehalose synthesis during starvation to support survival. Elife 2017, 6, e30057. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Chandra, A.; Mitic, L.L.; Onken, B.; Driscoll, M.; Kenyon, C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008, 4, e24. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Gibney, P.A. Dietary Trehalose as a Bioactive Nutrient. Nutrients 2023, 15, 1393. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiri, T.J.; Viau, C.; Gu, X.; Xu, L.; Lu, Y.; Xia, J. The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans. Metabolites 2023, 13, 953. https://doi.org/10.3390/metabo13080953
Shiri TJ, Viau C, Gu X, Xu L, Lu Y, Xia J. The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans. Metabolites. 2023; 13(8):953. https://doi.org/10.3390/metabo13080953
Chicago/Turabian StyleShiri, Tanisha Jean, Charles Viau, Xue Gu, Lei Xu, Yao Lu, and Jianguo Xia. 2023. "The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans" Metabolites 13, no. 8: 953. https://doi.org/10.3390/metabo13080953
APA StyleShiri, T. J., Viau, C., Gu, X., Xu, L., Lu, Y., & Xia, J. (2023). The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans. Metabolites, 13(8), 953. https://doi.org/10.3390/metabo13080953