Liver and Muscle Transcriptomes Differ in Mid-Lactation Cows Divergent in Feed Efficiency in the Presence or Absence of Supplemental Rumen-Protected Choline
Abstract
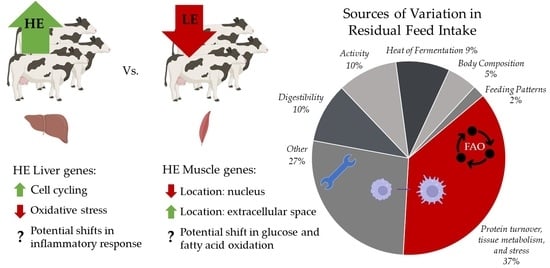
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Analysis
2.1.1. RFI Calculation
2.1.2. Blood Sample Collection and Metabolite Quantification
2.1.3. Liver and Muscle Tissue Collection and Analysis
2.2. Statistics
3. Results
3.1. High and Low RFI Grouping
3.2. Production Response of RPC Supplementation
3.3. Blood Metabolites
3.4. Liver and Muscle Tissue Transcriptome
4. Discussion
4.1. Response of RPC Supplementation
4.2. Blood Metabolite and Tissue Difference by RFI
4.3. Interaction of RFI Group and TRT on Blood Metabolites
4.4. Liver and Muscle Transcriptome by RFI Group
4.4.1. Liver Transcriptome
4.4.2. Muscle Transcriptome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- US Department of Agriculture-Economic Research Service Milk Cost of Production Estimates. Available online: http://www.ers.usda.gov/data-products/milk-cost-of-production-estimates/ (accessed on 12 May 2023).
- Koch, R.M.; Swiger, L.A.; Chambers, D.; Gregory, K.E. Efficiency of Feed Use in Beef Cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- VandeHaar, M.J.; Armentano, L.E.; Weigel, K.; Spurlock, D.M.; Tempelman, R.J.; Veerkamp, R. Harnessing the Genetics of the Modern Dairy Cow to Continue Improvements in Feed Efficiency. J. Dairy Sci. 2016, 99, 4941–4954. [Google Scholar] [CrossRef]
- Li, B.; Vanraden, P.M.; Guduk, E.; O’Connell, J.R.; Null, D.J.; Connor, E.E.; VandeHaar, M.J.; Tempelman, R.J.; Weigel, K.A.; Cole, J.B. Genomic Prediction of Residual Feed Intake in US Holstein Dairy Cattle. J. Dairy Sci. 2020, 103, 2477–2486. [Google Scholar] [CrossRef]
- Manzanilla-Pech, C.I.V.; Veerkamp, R.F.; Tempelman, R.J.; van Pelt, M.L.; Weigel, K.A.; VandeHaar, M.; Lawlor, T.J.; Spurlock, D.M.; Armentano, L.E.; Staples, C.R.; et al. Genetic Parameters between Feed-Intake-Related Traits and Conformation in 2 Separate Dairy Populations-the Netherlands and United States. J. Dairy Sci. 2016, 99, 443–457. [Google Scholar] [CrossRef]
- Tempelman, R.J.; Spurlock, D.M.; Coffey, M.; Veerkamp, R.F.; Armentano, L.E.; Weigel, K.A.; de Haas, Y.; Staples, C.R.; Connor, E.E.; Lu, Y.; et al. Heterogeneity in Genetic and Nongenetic Variation and Energy Sink Relationships for Residual Feed Intake across Research Stations and Countries. J. Dairy Sci. 2015, 98, 2013–2026. [Google Scholar] [CrossRef]
- VanRaden, P.; O’Connell, J.; Connor, E.; VandeHaar, M.; Tempelman, R.; Weigel, K. Including Feed Intake Data from U.S. Holsteins in Genomic Prediction. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Vol. Biology–Feed Intake and Efficiency 1. World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 11–16 February 2018. [Google Scholar]
- VanRaden, P.M.; Cole, J.B.; Parker Gaddis, K.L. Net Merit as a Measure of Lifetime Profit: 2021 Revision; AIP Reserach Reports; United States Department of Agriculture: Washington, DC, USA, 2021; pp. 1–20. [Google Scholar]
- Herd, R.M.; Arthur, P.F. Physiological Basis for Residual Feed Intake. J. Anim. Sci. 2009, 87, 64–71. [Google Scholar] [CrossRef]
- Martin, M.J.; Pralle, R.S.; Bernstein, I.R.; Vandehaar, M.J.; Weigel, K.A.; Zhou, Z.; White, H.M. Circulating Metabolites Indicate Differences in High and Low Residual Feed Intake Holstein Dairy Cows. Metabolites 2021, 11, 868. [Google Scholar] [CrossRef]
- Kennedy, K.M.; Becker, F.; Hammon, H.M.; Kuhla, B. Differences in Net Fat Oxidation, Heat Production, and Liver Mitochondrial DNA Copy Numbers between High and Low Feed-Efficient Dairy Cows. J. Dairy Sci. 2021, 104, 9287–9303. [Google Scholar] [CrossRef]
- Arshad, U.; Zenobi, M.G.; Staples, C.R.; Santos, J.E.P. Meta-Analysis of the Effects of Supplemental Rumen-Protected Choline during the Transition Period on Performance and Health of Parous Dairy Cows. J. Dairy Sci. 2020, 103, 282–300. [Google Scholar] [CrossRef]
- Chandler, T.L.; White, H.M. Choline and Methionine Differentially Alter Methyl Carbon Metabolism in Bovine Neonatal Hepatocytes. PLoS ONE 2017, 12, e0171080. [Google Scholar] [CrossRef]
- Sun, F.; Cao, Y.; Cai, C.; Li, S.; Yu, C.; Yao, J. Regulation of Nutritional Metabolism in Transition Dairy Cows: Energy Homeostasis and Health in Response to Post-Ruminal Choline and Methionine. PLoS ONE 2016, 11, e0160659. [Google Scholar] [CrossRef]
- Chandler, T.L.; White, H.M. Glucose Metabolism Is Differentially Altered by Choline and Methionine in Bovine Neonatal Hepatocytes. PLoS ONE 2019, 14, e0217160. [Google Scholar] [CrossRef] [PubMed]
- Piepenbrink, M.S.; Overton, T.R. Liver Metabolism and Production of Cows Fed Increasing Amounts of Rumen-Protected Choline during the Periparturient Period. J. Dairy Sci. 2003, 86, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Zenobi, M.G.; Scheffler, T.L.; Zuniga, J.E.; Poindexter, M.B.; Campagna, S.R.; Castro Gonzalez, H.F.; Farmer, A.T.; Barton, B.A.; Santos, J.E.P.; Staples, C.R. Feeding Increasing Amounts of Ruminally Protected Choline Decreased Fatty Liver in Nonlactating, Pregnant Holstein Cows in Negative Energy Status. J. Dairy Sci. 2018, 101, 5902–5923. [Google Scholar] [CrossRef]
- Zenobi, M.G.; Gardinal, R.; Zuniga, J.E.; Mamedova, L.K.; Driver, J.P.; Barton, B.A.; Santos, J.E.P.; Staples, C.R.; Nelson, C.D. Effect of Prepartum Energy Intake and Supplementation with Ruminally Protected Choline on Innate and Adaptive Immunity of Multiparous Holstein Cows. J. Dairy Sci. 2020, 103, 2200–2216. [Google Scholar] [CrossRef]
- Connor, E.E.; Hutchison, J.L.; Van Tassell, C.P.; Cole, J.B. Defining the Optimal Period Length and Stage of Growth or Lactation to Estimate Residual Feed Intake in Dairy Cows. J. Dairy Sci. 2019, 102, 6131–6143. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Dairy Cattle, 7th ed.; The National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- National Academies of Sciences; Engineering; Medicine. Nutrient Requirements of Dairy Cattle: Eighth Revised Edition; The National Academies Press: Washington, DC, USA, 2021; ISBN 978-0-309-67777-6. [Google Scholar]
- Shreve, B.; Thiex, N.; Wolf, M. National Forage Testing Association Reference Method: Dry Matter by Oven Drying for 3 Hours at 105 C; NFTA Reference Methods; National Forage Testing Association: Omaha, NE, USA, 2006. [Google Scholar]
- AOAC International. Official Methods of Analysis, 19th ed.; AOAC International: Gaithersburg, MD, USA, 2012. [Google Scholar]
- AOAC International. Official Methods of Analysis, 18th ed.; AOAC International: Arlington, VA, USA, 2005. [Google Scholar]
- AOAC International. Official Methods of Analysis, 15th ed.; AOAC International: Arlington, VA, USA, 1996. [Google Scholar]
- Derias, R. Method for Determination Water Soluble Carbohydrates. J. Sci. Food Agric. 1961, 12, 152. [Google Scholar]
- Bach Knudsen, K.E. Carbohydrate and Lignin Contents of Plant Materials Used in Animal Feeding. Anim. Feed Sci. Technol. 1997, 67, 319–338. [Google Scholar] [CrossRef]
- Dairy Record Managment Services. DHI Glossary; DRMS: Ames, IA, USA, 2010. [Google Scholar]
- Edmonson, A.J.; Lean, I.J.; Weaver, L.D.; Farver, T.; Webster, G. A Body Condition Scoring Chart for Holstein Dairy Cows. J. Dairy Sci. 1989, 72, 68–78. [Google Scholar] [CrossRef]
- Holtenius, P.; Holtenius, K. A Model to Estimate Insulin Sensitivity in Dairy Cows. Acta Vet. Scand. 2007, 49, 29. [Google Scholar] [CrossRef]
- Abuelo, Á.; De Koster, J.; Hernández, J.; Opsomer, G.; Grufman, L.; Castillo, C. Quantifying Bovine Insulin: Conversion of Units. Vet. Clin. Pathol. 2012, 41, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Sukhija, P.S.; Palmquist, D.L. Rapid Method for Determination of Total Fatty Acid Content and Composition of Feedstuffs and Feces. J. Agric. Food Chem. 1988, 36, 1202–1206. [Google Scholar] [CrossRef]
- Lucy, M.C.; Verkerk, G.A.; Whyte, B.E.; Macdonald, K.A.; Burton, L.; Cursons, R.T.; Roche, J.R.; Holmes, C.W. Somatotropic Axis Components and Nutrient Partitioning in Genetically Diverse Dairy Cows Managed under Different Feed Allowances in a Pasture System. J. Dairy Sci. 2009, 92, 526–539. [Google Scholar] [CrossRef]
- Roche, J.R.; Donkin, S.S.; Handley, R.R.; Heiser, A.; Walker, C.G.; Loor, J.J.; Mitchell, M.D.; Snell, R.G.; Henty, K.M.; White, H.M.; et al. Epigenetic Regulation of Pyruvate Carboxylase Gene Expression in the Postpartum Liver. J. Dairy Sci. 2016, 99, 5820–5827. [Google Scholar] [CrossRef]
- Pralle, R.S.; Erb, S.J.; Holdorf, H.T.; White, H.M. Greater Liver PNPLA3 Protein Abundance in Vivo and in Vitro Supports Lower Triglyceride Accumulation in Dairy Cows. Sci. Rep. 2021, 11, 2839. [Google Scholar] [CrossRef]
- Caputo Oliveira, R.; Erb, S.J.; Pralle, R.S.; Holdorf, H.T.; Seely, C.R.; White, H.M. Postpartum Supplementation with Fermented Ammoniated Condensed Whey Altered Nutrient Partitioning to Support Hepatic Metabolism. J. Dairy Sci. 2020, 103, 7055–7067. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P. A Reagent for the Single-Step Simultaneous Isolation of RNA, DNA and Proteins from Cell and Tissue Samples. Biotechniques 1993, 15, 532–534, 536–537. [Google Scholar] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression and Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Connor, E.E. Invited Review: Improving Feed Efficiency in Dairy Production: Challenges and Possibilities. Animal 2015, 9, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E. Symposium Review: Genomic Selection for Reducing Environmental Impact and Adapting to Climate Change. J. Dairy Sci. 2020, 103, 5366–5375. [Google Scholar] [CrossRef] [PubMed]
- Gaddis, K.; VanRaden, P.; Tempelman, R.; Weigel, K.; White, H.; Peñagaricano, F.; Koltes, J.; Santos, J.; Baldwin, R.; Burchard, J.; et al. Implementation of Feed Saved Evaluations in the U.S. Interbull Bull. 2021, 56, 147–152. [Google Scholar]
- Zeisel, S.H.; Da Costa, K.A. Choline: An Essential Nutrient for Public Health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef]
- Nutrient Research Council. Nutrient Requirements of Poultry, 9th ed.; The National Academies Press: Washington, DC, USA, 1994; Volume 42. [Google Scholar]
- Artegoitia, V.M.; Middleton, J.L.; Harte, F.M.; Campagna, S.R.; De Veth, M.J. Choline and Choline Metabolite Patterns and Associations in Blood and Milk during Lactation in Dairy Cows. PLoS ONE 2014, 9, e103412. [Google Scholar] [CrossRef]
- Pinotti, L.; Baldi, A.; Dell’Orto, V. Comparative Mammalian Choline Metabolism with Emphasis on the High-Yielding Dairy Cow. Nutr. Res. Rev. 2002, 15, 315–332. [Google Scholar] [CrossRef]
- Bauman, D.E.; Currie, W.B. Partitioning of Nutrients during Pregnancy and Lactation: A Review of Mechanisms Involving Homeostasis and Homeorhesis. J. Dairy Sci. 1980, 63, 1514–1529. [Google Scholar] [CrossRef]
- McFadden, J.W.; Girard, C.L.; Tao, S.; Zhou, Z.; Bernard, J.K.; Duplessis, M.; White, H.M. Symposium Review: One-Carbon Metabolism and Methyl Donor Nutrition in the Dairy Cow. J. Dairy Sci. 2020, 103, 5668–5683. [Google Scholar] [CrossRef]
- Mohsen, M.K.; Gaafar, H.M.A.; Shitta, A.A.; Yousif, A.M. Effect of Rumen Protected Choline Supplementation on Digestibility, Rumen Activity and Milk Yield in Lactating Friesian Cows. Slovak J. Anim. Sci. 2011, 44, 2011–2013. [Google Scholar]
- Davidson, S.; Hopkins, B.A.; Odle, J.; Brownie, C.; Fellner, V.; Whitlow, L.W. Supplementing Limited Methionine Diets with Rumen-Protected Methionine, Betaine, and Choline in Early Lactation Holstein Cows. J. Dairy Sci. 2008, 91, 1552–1559. [Google Scholar] [CrossRef] [PubMed]
- Erdman, R.A.; Sharma, B.K. Effect of Dietary Rumen-Protected Choline in Lactating Dairy Cows. J. Dairy Sci. 1991, 74, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Deuchler, K.N.; Piperova, L.S.; Erdman, R.A. Milk Choline Secretion as an Indirect Indicator of Postruminal Choline Supply. J. Dairy Sci. 1998, 81, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Bollatti, J.M.; Zenobi, M.G.; Artusso, N.A.; Alfaro, G.F.; Lopez, A.M.; Barton, B.A.; Nelson, C.D.; Staples, C.R.; Santos, J.E.P. Timing of Initiation and Duration of Feeding Rumen-Protected Choline Affects Performance of Lactating Holstein Cows. J. Dairy Sci. 2020, 103, 4174–4191. [Google Scholar] [CrossRef]
- Holdorf, H.T.; White, H.M. Effects of Rumen-Protected Choline Supplementation in Holstein Dairy Cows during Electric Heat Blanket-Induced Heat Stress. J. Dairy Sci. 2021, 104, 9715–9725. [Google Scholar] [CrossRef]
- Bertoni, G.; Trevisi, E.; Han, X.; Bionaz, M. Effects of Inflammatory Conditions on Liver Activity in Puerperium Period and Consequences for Performance in Dairy Cows. J. Dairy Sci. 2008, 91, 3300–3310. [Google Scholar] [CrossRef]
- Rahmani, M.; Dehghan-Banadaky, M.; Kamalyan, R. Comparison between Feeding Rumen-Protected Choline and Vitamin e on Milk Yield and Blood Metabolites in Early Lactation Dairy Cows. Anim. Prod. Sci. 2015, 55, 752–757. [Google Scholar] [CrossRef]
- Salam Karim, Y.; Hachim, S.K.; Abdul Ali, A.; Ameen Baqer, A.; Ali Yaseen, M.; Lafta, H.A.; Hussein Adhab, Z.; Ayad Kareem, H.; Shaker Hamza, I.; Hamad, D.A. The Influence of Rumen-Protected Choline and α-Tocopherol Supplementation on Early Lactating Dairy Cows Metabolism. Arch. Razi Inst. 2022, 77, 1165–1171. [Google Scholar] [CrossRef]
- Cook, H.W.; McMaster, C.R. Fatty Acid Desaturation and Chain Elongation in Eukaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes; Vance, D.E., Vance, F.F., Eds.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2002; Volume 36, pp. 181–204. ISBN 0444511385. [Google Scholar]
- Brash, A.R. Arachidonic Acid as a Bioactive Molecule. J. Clin. Investig. 2001, 107, 1339–1345. [Google Scholar] [CrossRef]
- Rodwell, V.W.; Bender, D.A.; Botham, K.M.; Kennelly, P.J.; Weil, P.A. Harper’s Illustrated Biochemestry, 30th ed.; McGraw Hill Professional: New York, NY, USA, 2015; ISBN 3527312188. [Google Scholar]
- Bergman, E.N. Hyperketonemia-Ketogenesis and Ketone Body Metabolism. J. Dairy Sci. 1971, 54, 936–948. [Google Scholar] [CrossRef]
- McArt, J.A.A.; Nydam, D.V.; Oetzel, G.R. Epidemiology of Subclinical Ketosis in Early Lactation Dairy Cattle. J. Dairy Sci. 2012, 95, 5056–5066. [Google Scholar] [CrossRef] [PubMed]
- Nehme Marinho, M.; Santos, J.E.P. Association of Residual Feed Intake with Blood Metabolites and Reproduction in Holstein Cows. Front. Anim. Sci. 2022, 3, 847574. [Google Scholar] [CrossRef]
- Martin, M.J.; Weigel, K.A.; White, H.M. Assessment of the Relationship between Postpartum Health and Mid-Lactation Performance, Behavior, and Feed Efficiency in Holstein Dairy Cows. Animals 2021, 11, 1385. [Google Scholar] [CrossRef] [PubMed]
- Nehme Marinho, M.; Zimpel, R.; Peñagaricano, F.; Santos, J.E.P. Assessing Feed Efficiency in Early and Mid Lactation and Its Associations with Performance and Health in Holstein Cows. J. Dairy Sci. 2021, 104, 5493–5507. [Google Scholar] [CrossRef]
- Rathbun, F.M.; Pralle, R.S.; Bertics, S.J.; Armentano, L.E.; Cho, K.; Do, C.; Weigel, K.A.; White, H.M. Relationships between Body Condition Score Change, Prior Mid-Lactation Phenotypic Residual Feed Intake, and Hyperketonemia Onset in Transition Dairy Cows. J. Dairy Sci. 2017, 100, 3685–3696. [Google Scholar] [CrossRef]
- Xu, G.; Ye, J.; Liu, J.; Yu, Y. Effect of Rumen-Protected Choline Addition on Milk Performance and Blood Metabolic Parameters in Transition Dairy Cows. Asian-Australas. J. Anim. Sci. 2006, 19, 390–395. [Google Scholar] [CrossRef]
- Zhou, Z.; Vailati-Riboni, M.; Trevisi, E.; Drackley, J.K.; Luchini, D.N.; Loor, J.J. Better Postpartal Performance in Dairy Cows Supplemented with Rumen-Protected Methionine Compared with Choline during the Peripartal Period. J. Dairy Sci. 2016, 99, 8716–8732. [Google Scholar] [CrossRef]
- Rigout, S.; Lemosquet, S.; Van Eys, J.E.; Blum, J.W.; Rulquin, H. Duodenal Glucose Increases Glucose Fluxes and Lactose Synthesis in Grass Silage-Fed Dairy Cows. J. Dairy Sci. 2002, 85, 595–606. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Kristensen, N.B.; Donkin, S.S.; Hammon, H.M.; Penner, G.B. Gluconeogenesis in Dairy Cows: The Secret of Making Sweet Milk from Sour Dough. IUBMB Life 2010, 62, 869–877. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, Z.; Wang, D.; Liu, J. Nitrogen Partitioning and Microbial Protein Synthesis in Lactating Dairy Cows with Different Phenotypic Residual Feed Intake. J. Anim. Sci. Biotechnol. 2019, 10, 54. [Google Scholar] [CrossRef]
- Du, X.; Chen, L.; Huang, D.; Peng, Z.; Zhao, C.; Zhang, Y.; Zhu, Y.; Wang, Z.; Li, X.; Liu, G. Elevated Apoptosis in the Liver of Dairy Cows with Ketosis. Cell. Physiol. Biochem. 2017, 43, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ding, H.Y.; Wang, X.C.; Feng, S.B.; Li, X.B.; Wang, Z.; Liu, G.W.; Li, X.W. An Association between the Level of Oxidative Stress and the Concentrations of NEFA and BHBA in the Plasma of Ketotic Dairy Cows. J. Anim. Physiol. Anim. Nutr. 2016, 100, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kadarmideen, H.N. Metabolomics Analyses in High-Low Feed Efficient Dairy Cows Reveal Novel Biochemical Mechanisms and Predictive Biomarkers. Metabolites 2019, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Vlaeminck, B.; Fievez, V.; Cabrita, A.R.J.; Fonseca, A.J.M.; Dewhurst, R.J. Factors Affecting Odd- and Branched-Chain Fatty Acids in Milk: A Review. Anim. Feed Sci. Technol. 2006, 131, 389–417. [Google Scholar] [CrossRef]
- Dodds, P.F.; Guzman, M.G.; Chalberg, S.C.; Anderson, G.J.; Kumar, S. Acetoacetyl-CoA Reductase Activity of Lactating Bovine Mammary Fatty Acid Synthase. J. Biol. Chem. 1981, 256, 6282–6290. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Gervais, R.; Rahman, M.M.; Gadeyne, F.; Gorniak, M.; Doreau, M.; Fievez, V. Postruminal Synthesis Modifies the Odd- and Branched-Chain Fatty Acid Profile from the Duodenum to Milk. J. Dairy Sci. 2015, 98, 4829–4840. [Google Scholar] [CrossRef]
- Shackelford, R.E.; Kaufmann, W.K.; Paules, R.S. Oxidative Stress and Cell Cycle Checkpoint Function. Free Radic. Biol. Med. 2000, 28, 1387–1404. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The P53 Pathway: Positive and Negative Feedback Loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- Grubbs, J.K.; Fritchen, A.N.; Huff-Lonergan, E.; Gabler, N.K.; Lonergan, S.M. Selection for Residual Feed Intake Alters the Mitochondria Protein Profile in Pigs. J. Proteom. 2013, 80, 334–345. [Google Scholar] [CrossRef]
- Ojano-Dirain, C.; Iqbal, M.; Wing, T.; Cooper, M.; Bottje, W. Glutathione and Respiratory Chain Complex Activity in Duodenal Mitochondria of Broilers with Low and High Feed Efficiency. Poult. Sci. 2005, 84, 782–788. [Google Scholar] [CrossRef]
- Iqbal, M.; Pumford, N.R.; Tang, Z.X.; Lassiter, K.; Ojano-Dirain, C.; Wing, T.; Cooper, M.; Bottje, W. Compromised Liver Mitochondrial Function and Complex Activity in Low Feed Efficient Broilers Are Associated with Higher Oxidative Stress and Differential Protein Expression. Poult. Sci. 2005, 84, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Casal, A.; Garcia-Roche, M.; Navajas, E.A.; Cassina, A.; Carriquiry, M. Differential Hepatic Oxidative Status in Steers with Divergent Residual Feed Intake Phenotype. Animal 2020, 14, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Davies, K.J.A. Oxidative DNA Damage & Repair: An Introduction. Free. Radic. Biol. Med. 2017, 107, 2–12. [Google Scholar] [CrossRef]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, Oxidants, and Aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial Dysfunction and Oxidative Stress in Metabolic Disorders—A Step towards Mitochondria Based Therapeutic Strategies. Biochim. Biophys. Acta-Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Bottje, W.; Tang, Z.X.; Iqbal, M.; Cawthon, D.; Okimoto, R.; Wing, T.; Cooper, M. Association of Mitochondrial Function with Feed Efficiency within a Single Genetic Line of Male Broilers. Poult. Sci. 2002, 81, 546–555. [Google Scholar] [CrossRef]
- Kolath, W.H.; Kerley, M.S.; Golden, J.W.; Keisler, D.H. The Relationship between Mitochondrial Function and Residual Feed Intake in Angus Steers. J. Anim. Sci. 2006, 84, 861–865. [Google Scholar] [CrossRef]
- Lancaster, P.A.; Carstens, G.E.; Michal, J.J.; Brennan, K.M.; Johnson, K.A.; Davis, M.E. Relationships between Residual Feed Intake and Hepatic Mitochondrial Function in Growing Beef Cattle. J. Anim. Sci. 2014, 92, 3134–3141. [Google Scholar] [CrossRef]
- Munford, R.S.; Weiss, J.P.; Lu, M. Biochemical Transformation of Bacterial Lipopolysaccharides by Acyloxyacyl Hydrolase Reduces Host Injury and Promotes Recovery. J. Biol. Chem. 2020, 295, 17842–17851. [Google Scholar] [CrossRef]
- Zou, B.; Jiang, W.; Han, H.; Li, J.; Mao, W.; Tang, Z.; Yang, Q.; Qian, G.; Qian, J.; Zeng, W.; et al. Acyloxyacyl Hydrolase Promotes the Resolution of Lipopolysaccharide-Induced Acute Lung Injury. PLoS Pathog. 2017, 13, e1006436. [Google Scholar] [CrossRef]
- Elchaninov, A.; Lokhonina, A.; Vishnyakova, P.; Soboleva, A.; Poltavets, A.; Artemova, D.; Makarov, A.; Glinkina, V.; Goldshtein, D.; Bolshakova, G.; et al. Marco+ Macrophage Dynamics in Regenerating Liver after 70% Liver Resection in Mice. Biomedicines 2021, 9, 1129. [Google Scholar] [CrossRef] [PubMed]
- Keshav, S.; Chung, P.; Milon, G.; Gordon, S. Lysozyme Is an Inducible Marker of Macrophage Activation in Murine Tissues as Demonstrated by in Situ Hybridization. J. Exp. Med. 1991, 174, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Dambuza, I.M.; Brown, G.D. C-Type Lectins in Immunity: Recent Developments. Curr. Opin. Immunol. 2015, 32, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.M.; Inamine, T.; Hochrath, K.; Chen, P.; Wang, L.; Llorente, C.; Bluemel, S.; Hartmann, P.; Xu, J.; Koyama, Y.; et al. Intestinal Fungi Contribute to Development of Alcoholic Liver Disease. J. Clin. Investig. 2017, 127, 2829–2841. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, J.R.; Boddicker, N.J.; Serão, N.V.L.; Young, J.M.; Rowland, R.R.R.; Dekkers, J.C.M. Response of Pigs Divergently Selected for Residual Feed Intake to Experimental Infection with the PRRS Virus. Livest. Sci. 2015, 177, 132–141. [Google Scholar] [CrossRef]
- Cheema, M.A.; Qureshi, M.A.; Havenstein, G.B. A Comparison of the Immune Response of a 2001 Commercial Broiler with a 1957 Randombred Broiler Strain When Fed Representative 1957 and 2001 Broiler Diets. Poult. Sci. 2003, 82, 1519–1529. [Google Scholar] [CrossRef]
- Hou, Y.; Bickhart, D.M.; Chung, H.; Hutchison, J.L.; Norman, H.D.; Connor, E.E.; Liu, G.E. Analysis of Copy Number Variations in Holstein Cows Identify Potential Mechanisms Contributing to Differences in Residual Feed Intake. Funct. Integr. Genom. 2012, 12, 717–723. [Google Scholar] [CrossRef]
- Kvidera, S.K.; Horst, E.A.; Abuajamieh, M.; Mayorga, E.J.; Fernandez, M.V.S.; Baumgard, L.H. Glucose Requirements of an Activated Immune System in Lactating Holstein Cows. J. Dairy Sci. 2017, 100, 2360–2374. [Google Scholar] [CrossRef]
- Shattuck-Heidorn, H.; Reiches, M.W.; Prentice, A.M.; Moore, S.E.; Ellison, P.T. Energetics and the Immune System: Trade-Offs Associated with Non-Acute Levels of CRP in Adolescent Gambian Girls. Evol. Med. Public Health 2017, 2017, 27–38. [Google Scholar] [CrossRef]
- Ganeshan, K.; Nikkanen, J.; Man, K.; Leong, Y.A.; Sogawa, Y.; Maschek, J.A.; Van Ry, T.; Chagwedera, D.N.; Cox, J.E.; Chawla, A. Energetic Trade-Offs and Hypometabolic States Promote Disease Tolerance. Cell 2019, 177, 399–413.e12. [Google Scholar] [CrossRef]
- Menikdiwela, K.R.; Ramalingam, L.; Abbas, M.M.; Bensmail, H.; Scoggin, S.; Kalupahana, N.S.; Palat, A.; Gunaratne, P.; Moustaid-Moussa, N. Role of MicroRNA 690 in Mediating Angiotensin II Effects on Inflammation and Endoplasmic Reticulum Stress. Cells 2020, 9, 1327. [Google Scholar] [CrossRef] [PubMed]
- Fujishiro, M.; Gotoh, Y.; Katagiri, H.; Sakoda, H.; Ogihara, T.; Anai, M.; Onishi, Y.; Ono, H.; Funaki, M.; Inukai, K.; et al. MKK6/3 and P38 MAPK Pathway Activation Is Not Necessary for Insulin-Induced Glucose Uptake but Regulates Glucose Transporter Expression. J. Biol. Chem. 2001, 276, 19800–19806. [Google Scholar] [CrossRef] [PubMed]
- Ntambi, J.M.; Miyazaki, M. Regulation of Stearoyl-CoA Desaturases and Role in Metabolism. Prog. Lipid Res. 2004, 43, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional Lipid Metabolism Proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Hotamisligil, G.S. Fatty Acid-Binding Proteins: Role in Metabolic Diseases and Potential as Drug Targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Feige, J.N.; Gelman, L.; Michalik, L.; Desvergne, B.; Wahli, W. From Molecular Action to Physiological Outputs: Peroxisome Proliferator-Activated Receptors Are Nuclear Receptors at the Crossroads of Key Cellular Functions. Prog. Lipid Res. 2006, 45, 120–159. [Google Scholar] [CrossRef]
- Collin de I’Hortet, A.; Takeishi, K.; Guzman-lepe, J.; Morita, K.; Achreja, A.; Popovic, B.; Wang, Y.; Handa, K.; Mittal, A.; Meurs, N.; et al. Generation of Human Fatty Livers Using Custom-Engineered Induced Pluripotent Stem Cells with Modifiable SIRT1 Metabolism. Cell Metab. 2019, 30, 385–401. [Google Scholar] [CrossRef]
- Lim, J.H.; Gerhart-Hines, Z.; Dominy, J.E.; Lee, Y.; Kim, S.; Tabata, M.; Xiang, Y.K.; Puigserver, P. Oleic Acid Stimulates Complete Oxidation of Fatty Acids through Protein Kinase A-Dependent Activation of SIRT1-PGC1α Complex. J. Biol. Chem. 2013, 288, 7117–7126. [Google Scholar] [CrossRef]
- Erb, S.J.; Chandler, T.L.; White, H.M. Responsiveness of PNPLA3 and Lipid-Related Transcription Factors Is Dependent upon Fatty Acid Profile in Primary Bovine Hepatocytes. Sci. Rep. 2022, 12, 888. [Google Scholar] [CrossRef]
- Bedu, E.; Wahli, W.; Desvergne, B. Peroxisome Proliferator-Activated Receptor β/δ as a Therapeutic Target for Metabolic Diseases. Expert Opin. Ther. Targets 2005, 9, 861–873. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of Metabolism and Mitochondrial Homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]


| CTL | RPC | |||
|---|---|---|---|---|
| Item, %DM | Mean | SD | Mean | SD |
| Ingredient composition | ||||
| Alfalfa haylage | 23.62 | 0.73 | 23.87 | 1.05 |
| Corn silage | 29.92 | 0.97 | 30.03 | 1.25 |
| Distillers grain | 1.22 | 0.06 | 1.18 | 0.03 |
| Cottonseed | 5.09 | 0.49 | 5.04 | 0.37 |
| High moisture corn | 12.66 | 0.37 | 12.42 | 0.73 |
| Protein concentrate mix 1 | 27.49 | 0.62 | 27.46 | 0.70 |
| Nutrient analysis | ||||
| DM, % as fed | 50.04 | 49.94 | ||
| CP | 18.10 | 18.16 | ||
| aNDF | 28.07 | 27.65 | ||
| aNDFom | 27.28 | 26.85 | ||
| ADF | 19.98 | 20.07 | ||
| Lignin | 3.40 | 3.37 | ||
| NFC | 43.73 | 43.97 | ||
| Starch | 25.66 | 25.79 | ||
| Fat | 4.55 | 4.52 | ||
| NEL 3×, Mcal/kg DM 2 | 1.71 | 1.71 | ||
| Variable 1 | HE | LE | p-Value | ||
|---|---|---|---|---|---|
| RFI, kg | −1.73 | [−2.16, −1.30] | 1.72 | [1.17, 2.27] | - |
| DMI, kg | 29.5 | [28.5, 30.6] | 33.5 | [32.5, 34.6] | <0.01 |
| Milk Production | |||||
| Milk, kg | 50.7 | [46.7, 54.8] | 51.2 | [47.2, 55.2] | 0.87 |
| Milk energy, Mcal | 33.4 | [30.9, 36.0] | 34.4 | [31.8, 37.0] | 0.59 |
| ECM, kg | 49.7 | [46.0, 53.3] | 51.0 | [47.3, 54.6] | 0.61 |
| FCM, kg | 48.4 | [44.7, 52.2] | 50.0 | [46.3, 53.7] | 0.54 |
| Fat, kg | 2.7 | [2.2, 3.2] | 3.0 | [2.5, 3.5] | 0.32 |
| Protein, kg | 1.6 | [1.4, 1.7] | 1.6 | [1.4, 1.7] | 0.94 |
| Lactose, kg | 2.4 | [2.2, 2.6] | 2.4 | [2.2, 2.6] | 0.92 |
| Fat, % | 3.20 | [2.95, 3.45] | 3.37 | [3.12, 3.62] | 0.32 |
| Protein, % | 3.06 | [2.94, 3.17] | 3.06 | [2.94, 3.17] | 0.98 |
| MUN, mg/dL | 14.6 | [13.7, 15.6] | 15.3 | [14.4, 16.2] | 0.32 |
| Body Size | |||||
| BW, kg | 768 | [733, 802] | 784 | [750, 819] | 0.49 |
| Metabolic BW | 146 | [141, 151] | 148 | [143, 153] | 0.50 |
| BW change, kg/d | 0.32 | [0.12, 0.53] | 0.26 | [0.06, 0.47] | 0.69 |
| BCS | 3.22 | [3.07, 3.37] | 3.15 | [3.00, 3.30] | 0.48 |
| Variable 1 | CTL | RPC | p-Value | ||
|---|---|---|---|---|---|
| RFI, kg | −0.05 | [−0.53, 0.43] | 0.05 | [−0.42, 0.52] | 0.76 |
| DMI, kg | 31.5 | [30.7, 32.3] | 31.1 | [30.3, 31.9] | 0.47 |
| Milk Production | |||||
| Milk, kg | 51.9 | [49.6, 54.1] | 50.1 | [47.9, 52.2] | 0.25 |
| Milk energy, Mcal | 34.0 | [32.6, 35.4] | 33.6 | [32.2, 35.0] | 0.65 |
| ECM, kg | 50.6 | [48.6, 52.6] | 49.8 | [47.8, 51.8] | 0.58 |
| FCM, kg | 49.5 | [47.4, 51.6] | 48.6 | [46.5, 50.7] | 0.56 |
| Fat, kg | 1.66 | [1.57, 1.76] | 1.66 | [1.57, 1.75] | 0.95 |
| Protein, kg | 1.57 | [1.51, 1.64] | 1.56 | [1.50, 1.62] | 0.72 |
| Lactose, kg | 2.46 | [2.35, 2.57] | 2.38 | [2.27, 2.49] | 0.30 |
| Fat, % | 3.27 | [3.10, 3.43] | 3.40 | [3.24, 3.54] | 0.26 |
| Protein, % | 3.02 | [2.95, 3.08] | 3.09 | [3.03, 3.17] | 0.11 |
| MUN, mg/dL | 14.1 | [13.6, 14.5] | 14.7 | [14.2, 15.2] | 0.07 |
| Body Size | |||||
| BW, kg | 778 | [755, 802] | 759 | [736, 782] | 0.24 |
| Metabolic BW | 0.21 | [0.07, 0.36] | 0.14 | [0.00, 0.28] | 0.45 |
| BCS | 3.18 | [3.04, 3.31] | 3.13 | [3.00, 3.26] | 0.66 |
| HE 1 | LE 1 | p-Value 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite 3 | CTL | RPC | CTL | RPC | RFI | TRT | RxT | ||||
| BHB, mmol/L | 0.67 | [0.54, 0.83] | 0.75 | [0.60, 0.93] | 0.56 | [0.45, 0.70] | 0.62 | [0.50, 0.77] | 0.04 | 0.24 | 0.97 |
| Glucose, mg/dL | 63.3 ab | [60.6, 66.0] | 63.9 ab | [61.2, 66.6] | 62.8 b | [60.1, 65.6] | 67.8 a | [65.1, 70.4] | 0.19 | 0.03 | 0.09 |
| Fatty acids, mEq/L | 0.13 | [0.11, 0.16] | 0.12 | [0.09, 0.14] | 0.14 | [0.12, 0.17] | 0.14 | [0.11, 0.16] | 0.21 | 0.38 | 0.54 |
| Triglyceride, mg/dL | 9.9 a | [8.1, 11.7] | 7.5 b | [5.7, 9.4] | 9.4 ab | [7.1, 11.7] | 9.6 ab | [7.4, 11.8] | 0.25 | 0.12 | 0.05 |
| Albumin, g/dL | 3.73 | [3.62, 3.86] | 3.79 | [3.67, 3.93] | 3.67 | [3.57, 3.78] | 3.89 | [3.75, 4.06] | 0.84 | 0.03 | 0.20 |
| Bilirubin, mg/dL | 0.09 | [0.06, 0.12] | 0.11 | [0.04, 0.18] | 0.10 | [0.06, 0.14] | 0.08 | [0.07, 0.09] | 0.66 | 0.95 | 0.34 |
| BUN, mg/dL | 17.1 | [13.9, 20.3] | 17.8 | [14.6, 21.0] | 17.5 | [14.6, 20.4] | 18.3 | [15.5, 21.1] | 0.62 | 0.45 | 0.96 |
| Creatinine, mg/dL | 0.71 | [0.64, 0.78] | 0.69 | [0.62, 0.76] | 0.66 | [0.59, 0.73] | 0.71 | [0.64, 0.78] | 0.67 | 0.63 | 0.35 |
| ALT, U/L | 31.4 ab | [27.7, 35.1] | 24.7 b | [21, 28.4] | 33.0 a | [29.3, 36.7] | 36.0 a | [32.3, 39.6] | <0.01 | 0.31 | 0.01 |
| AST, U/L | 121 | [99, 143] | 126 | [103, 148] | 113 | [91, 136] | 133 | [111, 155] | 0.98 | 0.23 | 0.45 |
| Insulin, µg/L | 0.45 | [0.30, 0.69] | 0.60 | [0.39, 0.91] | 0.38 | [0.25, 0.58] | 0.38 | [0.25, 0.58] | 0.13 | 0.50 | 0.49 |
| RQUICKI 4 | 0.53 | [0.48, 0.59] | 0.54 | [0.40, 0.68] | 0.56 | [0.50, 0.62] | 0.54 | [0.48, 0.61] | 0.67 | 0.89 | 0.79 |
| HE 1 | LE 1 | p-Value 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fatty Acid, mg/dL 3 | CTL | RPC | CTL | RPC | RFI | TRT | RxT | ||||
| C14:0 | 0.42 | [0.38, 0.47] | 0.46 | [0.42, 0.51] | 0.46 | [0.42, 0.50] | 0.43 | [0.38, 0.47] | 0.95 | 0.81 | 0.08 |
| C15:0 | 0.31 | [0.28, 0.33] | 0.31 | [0.28, 0.33] | 0.32 | [0.29, 0.34] | 0.32 | [0.29, 0.34] | 0.31 | 0.94 | 0.94 |
| C16:0 | 4.10 | [3.63, 4.56] | 4.01 | [3.55, 4.47] | 4.00 | [3.53, 4.46] | 4.20 | [3.74, 4.67] | 0.84 | 0.79 | 0.52 |
| C16:1 | 0.69 | [0.61, 1.03] | 0.64 | [0.63, 0.69] | 0.67 | [0.63, 0.73] | 0.67 | [0.62, 0.74] | 0.85 | 0.91 | 0.17 |
| C17:0 | 0.87 | [0.78, 0.98] | 0.73 | [0.66, 0.80] | 0.80 | [0.72, 0.89] | 0.87 | [0.78, 0.99] | 0.29 | 0.36 | 0.02 |
| C18:0 | 4.72 | [4.16, 5.45] | 4.52 | [4.01, 5.19] | 4.64 | [4.10, 5.35] | 4.93 | [4.32, 5.73] | 0.59 | 0.91 | 0.43 |
| C18:1 | 3.15 | [2.84, 3.47] | 3.32 | [3.00, 3.63] | 3.35 | [3.03, 3.66] | 3.14 | [2.83, 3.46] | 0.94 | 0.90 | 0.24 |
| C18:2 | 13.87 | [11.8, 15.9] | 12.18 | [10.1, 14.2] | 13.63 | [11.5, 15.7] | 14.71 | [12.7, 16.8] | 0.24 | 0.75 | 0.16 |
| C18:3 | 2.21 | [1.88, 2.53] | 2.10 | [1.78, 2.43] | 2.30 | [1.98, 2.63] | 2.35 | [2.03, 2.67] | 0.28 | 0.86 | 0.63 |
| C20:3 | 1.39 | [1.33, 1.45] | 1.36 | [1.30, 1.41] | 1.40 | [1.25, 1.54] | 1.44 | [1.30, 1.59] | 0.38 | 0.91 | 0.43 |
| C20:4 | 0.73 | [0.69, 0.76] | 0.78 | [0.74, 0.82] | 0.73 | [0.70, 0.77] | 0.74 | [0.71, 0.78] | 0.37 | 0.09 | 0.21 |
| KEGG Pathway | ID | Genes | Fold Enrichment | Corrected p-Value | Gene Symbols 2 |
|---|---|---|---|---|---|
| Cell cycle | bta04110 | 14 | 8.4 | 6.9 ×10−12 | BUB1, BUB1B, CCNA2, CCNB1, CCNB2, CCNE2, CDC20, CDC25A, CDC6, CDK1, CDKN2C, ESPL1, ORC1, TTK |
| Cellular senescence | bta04218 | 8 | 4.8 | 0.00067 | CCNA2, CCNB1, CCNB2, CCNE2, CDC25A, CDK1, FOXM1, LOC787122 |
| Human T-cell leukemia virus 1 infection | bta05166 | 8 | 4.8 | 0.00540 | BUB1B, CCNA2, CCNB2, CCNE2, CDC20, CDKN2C, ESPL1, LOC787122 |
| p53 signaling pathway | bta04115 | 5 | 3.0 | 0.01500 | CCNB1, CCNB2, CCNE2, CDK1, RRM2 |
| Gene Ontology Domain | ID | Genes | Fold Enrichment | Corrected p-Value |
|---|---|---|---|---|
| Upregulated in HE | ||||
| Cell Component: extracellular space | GO:0005615 | 19 | 2.9 | 0.0130 |
| Molecular Function: actin filament binding | GO:0051015 | 8 | 5.3 | 0.0140 |
| Downregulated in HE | ||||
| Cell Component: nucleus | GO:0005634 | 44 | 1.9 | 0.0061 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, M.J.; Li, W.; Kendall, S.J.; Larsen, A.; Weigel, K.A.; White, H.M. Liver and Muscle Transcriptomes Differ in Mid-Lactation Cows Divergent in Feed Efficiency in the Presence or Absence of Supplemental Rumen-Protected Choline. Metabolites 2023, 13, 1023. https://doi.org/10.3390/metabo13091023
Caputo MJ, Li W, Kendall SJ, Larsen A, Weigel KA, White HM. Liver and Muscle Transcriptomes Differ in Mid-Lactation Cows Divergent in Feed Efficiency in the Presence or Absence of Supplemental Rumen-Protected Choline. Metabolites. 2023; 13(9):1023. https://doi.org/10.3390/metabo13091023
Chicago/Turabian StyleCaputo, Malia J., Wenli Li, Sophia J. Kendall, Anna Larsen, Kent A. Weigel, and Heather M. White. 2023. "Liver and Muscle Transcriptomes Differ in Mid-Lactation Cows Divergent in Feed Efficiency in the Presence or Absence of Supplemental Rumen-Protected Choline" Metabolites 13, no. 9: 1023. https://doi.org/10.3390/metabo13091023
APA StyleCaputo, M. J., Li, W., Kendall, S. J., Larsen, A., Weigel, K. A., & White, H. M. (2023). Liver and Muscle Transcriptomes Differ in Mid-Lactation Cows Divergent in Feed Efficiency in the Presence or Absence of Supplemental Rumen-Protected Choline. Metabolites, 13(9), 1023. https://doi.org/10.3390/metabo13091023