Integrative Multiomics Approach to Skin: The Sinergy between Individualised Medicine and Futuristic Precision Skin Care?
Abstract
:1. Introduction
2. Skin Microbiota
2.1. Composition
2.2. Role and Alteration
2.3. Manipulation
3. The Gut-Skin Brain Axis in Health and Disease
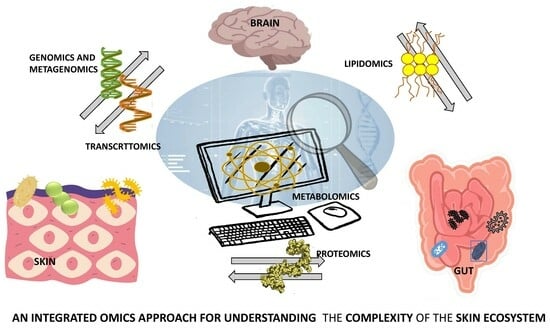
4. Integrative Multiomics Approach to Skin
5. The Future of Skin Care: Between Individualized Medicine and Precision Cosmetics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Moeller Christensen, G.J.; Nakatsuji, T.; et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, M. Skin Barrier Function and the Microbiome. Int. J. Mol. Sci. 2022, 23, 13071. [Google Scholar] [CrossRef]
- De Pessemier, B.; Grine, L.; Debaere, M.; Maes, A.; Paetzold, B.; Callewaert, C. Gut-Skin Axis: Current Knowledge of the Interrelationship between Microbial Dysbiosis and Skin Conditions. Microorganisms 2021, 9, 353. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Sandhu, S.S.; Pourang, A.; Sivamani, R.K. A review of next generation sequencing technologies used in the evaluation of the skin microbiome: What a time to be alive. Dermatol. Online J. 2019, 25, 1–8. [Google Scholar] [CrossRef]
- Jiang, B.; Jia, Y.; He, C. Promoting new concepts of skincare via skinomics and systems biology-From traditional skincare and efficacy-based skincare to precision skincare. J. Cosmet. Dermatol. 2018, 17, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 626. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Cassir, N.; Papazian, L.; Fournier, P.E.; Raoult, D.; La Scola, B. Insights into bacterial colonization of intensive care patients’ skin: The effect of chlorhexidine daily bathing. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 999–1004. [Google Scholar] [CrossRef]
- Myles, I.A.; Reckhow, J.D.; Williams, K.W.; Sastalla, I.; Frank, K.M.; Datta, S.K. A method for culturing Gram-negative skin microbiota. BMC Microbiol. 2016, 16, 60. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.F.; Oddos, T.; Stamatas, G. Deciphering the Role of Skin Surface Microbiome in Skin Health: An Integrative Multiomics Approach Reveals Three Distinct Metabolite–Microbe Clusters. J. Investig. Dermatol. 2022, 142, 469–479.e5. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mitra, R.; Maitra, A.; Gupta, S.; Kumaran, S.; Chakrabortty, A.; Majumder, P.P. Sebum and Hydration Levels in Specific Regions of Human Face Significantly Predict the Nature and Diversity of Facial Skin Microbiome. Sci. Rep. 2016, 6, 36062. [Google Scholar] [CrossRef]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 555, 543. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Scholz, C.F.P.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef]
- Grice, E.A. The intersection of microbiome and host at the skin interface: Genomic- and metagenomic-based insights. Genome Res. 2015, 25, 1514–1520. [Google Scholar] [CrossRef]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; NISC Comparative Sequencing Program; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Park, D.H.; Kim, J.W.; Park, H.J.; Hahm, D.H. Comparative Analysis of the Microbiome across the Gut-Skin Axis in Atopic Dermatitis. Int. J. Mol. Sci. 2021, 22, 4228. [Google Scholar] [CrossRef]
- Eigenmann, P.A.; Beyer, K.; Lack, G.; Muraro, A.; Ong, P.Y.; Sicherer, S.H.; Sampson, H.A. Are avoidance diets still warranted in children with atopic dermatitis? Pediatr. Allergy Immunol. 2020, 31, 19–26. [Google Scholar] [CrossRef]
- Dessì, A.; Di Maria, C.; Pintus, R.; Fanos, V.; Bosco, A. Lipidomics and Metabolomics in Infant Atopic Dermatitis: What’s the Correlation with Early Nutrition? Curr. Pediatr. Rev. 2023. [Google Scholar] [CrossRef]
- Callewaert, C.; Knödlseder, N.; Karoglan, A.; Güell, M.; Paetzold, B. Skin microbiome transplantation and manipulation: Current state of the art. Comput. Struct. Biotechnol. J. 2021, 19, 624–631. [Google Scholar] [CrossRef]
- Myles, I.A.; Earland, N.J.; Anderson, E.D.; Moore, I.N.; Kieh, M.D.; Williams, K.W.; Saleem, A.; Fontecilla, N.M.; Welch, P.A.; Darnell, D.A.; et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 2018, 3, e120608. [Google Scholar] [CrossRef]
- Paetzold, B.; Willis, J.R.; Pereira de Lima, J.; Knödlseder, N.; Brüggemann, H.; Quist, S.R.; Gabaldón, T.; Güell, M. Skin microbiome modulation induced by probiotic solutions. Microbiome 2019, 7, 95. [Google Scholar] [CrossRef]
- Mahmud, M.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Akter, S.; Hasan, M.R.; Acharjee, M.; Hossain, M.S.; et al. Impact of gut microbiome on skin health: Gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes 2022, 14, 2096995. [Google Scholar] [CrossRef]
- Polkowska-Pruszyńska, B.; Gerkowicz, A.; Krasowska, D. The gut microbiome alterations in allergic and inflammatory skin diseases—An update. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 455–464. [Google Scholar] [CrossRef]
- Jiao, Y.; Wu, L.; Huntington, N.D.; Zhang, X. Crosstalk Between Gut Microbiota and Innate Immunity and Its Implication in Autoimmune Diseases. Front. Immunol. 2020, 11, 282. [Google Scholar] [CrossRef]
- Lyte, M. Microbial endocrinology and the microbiota-gut-brain axis. Adv. Exp. Med. Biol. 2014, 817, 3–24. [Google Scholar]
- Bowe, W.P.; Logan, A.C. Acne vulgaris, probiotics and the gut-brain-skin axis—Back to the future? Gut Pathog. 2011, 3, 1. [Google Scholar] [CrossRef]
- Chen, G.; Chen, Z.M.; Fan, X.Y.; Jin, Y.L.; Li, X.; Wu, S.R.; Ge, W.W.; Lv, C.H.; Wang, Y.K.; Chen, J.G. Gut-Brain-Skin Axis in Psoriasis: A Review. Dermatol. Ther. 2021, 11, 25–38. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Kirjavainen, P.; Eerola, E.; Kero, P.; Salminen, S.; Isolauri, E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 2001, 107, 129–134. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; van den Brandt, P.A.; Kummeling, I.; Snijders, B.; Stelma, F.; Adams, H.; van Ree, R.; Stobberingh, E.E. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut 2007, 56, 661–667. [Google Scholar] [CrossRef]
- Watanabe, S.; Narisawa, Y.; Arase, S.; Okamatsu, H.; Ikenaga, T.; Tajiri, Y.; Kumemura, M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2003, 111, 587–591. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Song, H.; Yoo, Y.; Hwang, J.; Na, Y.C.; Kim, H.S. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol. 2016, 137, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Hokazono, H.; Omori, T.; Ono, K. Effects of single and combined administration of fermented barley extract and gamma-aminobutyric acid on the development of atopic dermatitis in NC/Nga mice. Biosci. Biotechnol. Biochem. 2010, 74, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Arndt, J.; Smith, N.; Tausk, F. Stress and atopic dermatitis. Curr. Allergy Asthma Rep. 2008, 8, 312–317. [Google Scholar] [CrossRef]
- Pondeljak, N.; Lugović-Mihić, L. Stress-induced Interaction of Skin Immune Cells, Hormones, and Neurotransmitters. Clin. Ther. 2020, 42, 757–770. [Google Scholar] [CrossRef]
- Tan, L.; Zhao, S.; Zhu, W.; Wu, L.; Li, J.; Shen, M.; Lei, L.; Chen, X.; Peng, C. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp. Dermatol. 2018, 27, 144–149. [Google Scholar] [CrossRef]
- Scher, J.U.; Ubeda, C.; Artacho, A.; Attur, M.; Isaac, S.; Reddy, S.M.; Marmon, S.; Neimann, A.; Brusca, S.; Patel, T.; et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015, 67, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A decade of metaproteomics: Where we stand and what the future holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef] [PubMed]
- Dessì, A.; Cesare Marincola, F.; Masili, A.; Gazzolo, D.; Fanos, V. Clinical metabolomics and nutrition: The new frontier in neonatology and pediatrics. Biomed. Res. Int. 2014, 2014, 981219. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Jia, Y. Application of omics technologies in dermatological research and skin management. J. Cosmet. Dermatol. 2022, 21, 451–460. [Google Scholar] [CrossRef]
- Brandão, L.A.C.; Tricarico, P.M.; Gratton, R.; Agrelli, A.; Zupin, L.; Abou-Saleh, H.; Moura, R.; Crovella, S. Multiomics Integration in Skin Diseases with Alterations in Notch Signaling Pathway: PlatOMICs Phase 1 Deployment. Int. J. Mol. Sci. 2021, 22, 1523. [Google Scholar] [CrossRef]
- Guo, Y.; Luo, L.; Zhu, J.; Li, C. Multi-Omics Research Strategies for Psoriasis and Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 8018. [Google Scholar] [CrossRef]
- Sun, Y.V.; Hu, Y.J. Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. 2016, 93, 147–190. [Google Scholar]
- Ashrafi, M.; Xu, Y.; Muhamadali, H.; White, I.; Wilkinson, M.; Hollywood, K.; Baguneid, M.; Goodacre, R.; Bayat, A. A microbiome and metabolomic signature of phases of cutaneous healing identified by profiling sequential acute wounds of human skin: An exploratory study. PLoS ONE 2020, 15, e0229545. [Google Scholar] [CrossRef]
- Emmert, H.; Baurecht, H.; Thielking, F.; Stölzl, D.; Rodriguez, E.; Harder, I.; Proksch, E.; Weidinger, S. Stratum corneum lipidomics analysis reveals altered ceramide profile in atopic dermatitis patients across body sites with correlated changes in skin microbiome. Exp. Dermatol. 2021, 30, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An integrative metabolomics and transcriptomics study to identify metabolic alterations in aged skin of humans in vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef]
- Marathe, S.; Dhamija, B.; Kumar, S.; Jain, N.; Ghosh, S.; Dharikar, J.P.; Srinivasan, S.; Das, S.; Sawant, A.; Desai, S.; et al. Multiomics Analysis and Systems Biology Integration Identifies the Roles of IL-9 in Keratinocyte Metabolic Reprogramming. J. Investig. Dermatol. 2021, 141, 1932–1942. [Google Scholar] [CrossRef]
- Acharjee, A.; Gribaleva, E.; Bano, S.; Gkoutos, G.V. Multi-omics-based identification of atopic dermatitis target genes and their potential associations with metabolites and miRNAs. Am. J. Transl. Res. 2021, 13, 13697–13709. [Google Scholar] [PubMed]
- Zhou, Y.; Wang, Y.; Su, J.; Wu, Z.; Wang, C.; Zhong, W.; Liu, X.; Cui, L.; Zhou, X.; Ma, Y.; et al. Integration of microRNAome, proteomics and metabolomics to analyze arsenic-induced malignant cell transformation. Oncotarget 2017, 8, 90879–90896. [Google Scholar] [CrossRef]
- Misra, N.; Clavaud, C.; Guinot, F.; Bourokba, N.; Nouveau, S.; Mezzache, S.; Palazzi, P.; Appenzeller, B.M.R.; Tenenhaus, A.; Leung, M.H.Y.; et al. Multi-omics analysis to decipher the molecular link between chronic exposure to pollution and human skin dysfunction. Sci. Rep. 2021, 11, 18302. [Google Scholar] [CrossRef] [PubMed]
- Tilton, S.C.; Matzke, M.M.; Sowa, M.B.; Stenoien, D.L.; Weber, T.J.; Morgan, W.F.; Waters, K.M. Data integration reveals key homeostatic mechanisms following low dose radiation exposure. Toxicol. Appl. Pharmacol. 2015, 285, 1–11. [Google Scholar] [CrossRef]
- Conwill, A.; Kuan, A.C.; Damerla, R.; Poret, A.J.; Baker, J.S.; Tripp, A.D.; Alm, E.J.; Lieberman, T.D. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 2022, 30, 171–182.e7. [Google Scholar] [CrossRef]
- Celebi Sozener, Z.; Ozdel Ozturk, B.; Cerci, P.; Turk, M.; Gorgulu Akin, B.; Akdis, M.; Altiner, S.; Ozbey, U.; Ogulur, I.; Mitamura, Y.; et al. Epithelial barrier hypothesis: Effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy 2022, 77, 1418–1449. [Google Scholar] [CrossRef]
- Martellacci, L.; Quaranta, G.; Patini, R.; Isola, G.; Gallenzi, P.; Masucci, L. A Literature Review of Metagenomics and Culturomics of the Peri-implant Microbiome: Current Evidence and Future Perspectives. Materials 2019, 12, 3010. [Google Scholar] [CrossRef]
- Katsnelson, A. Cosmetics: Molecular beauty. Nature 2015, 526, S4–S5. [Google Scholar] [CrossRef]
- Seité, S.; Zelenkova, H.; Martin, R. Clinical efficacy of emollients in atopic dermatitis patients—Relationship with the skin microbiota modification. Clin. Cosmet. Investig. Dermatol. 2017, 10, 25–33. [Google Scholar] [CrossRef]
- Yang, L.; Mao-Qiang, M.; Taljebini, M.; Elias, P.M.; Feingold, K.R. Topical stratum corneum lipids accelerate barrier repair after tape stripping, solvent treatment and some but not all types of detergent treatment. Br. J. Dermatol. 1995, 33, 679–685. [Google Scholar] [CrossRef]
- Sahle, F.F.; Gebre-Mariam, T.; Dobner, B.; Wohlrab, J.; Neubert, R.H. Skin diseases associated with the depletion of stratum corneum lipids and stratum corneum lipid substitution therapy. Skin. Pharmacol. Physiol. 2015, 28, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Touumazou, C.; Nedjai, B.; Sim, C. Product Selection Using Genetic Analysis. International Patent WO2013093407A1, 27 June 2013. [Google Scholar]
- Blumenberg, M. SKINOMICS: Transcriptional Profiling in Dermatology and Skin Biology. Curr. Genom. 2012, 13, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, A.; Righi, V.; Tarentini, E.; Magnoni, C. Current Knowledge in Skin Metabolomics: Updates from Literature Review. Int. J. Mol. Sci. 2022, 23, 8776. [Google Scholar] [CrossRef] [PubMed]
- Vineis, P.; Robinson, O.; Chadeau-Hyam, M.; Dehghan, A.; Mudway, I.; Dagnino, S. What is new in the exposome? Environ. Int. 2020, 143, 105887. [Google Scholar] [CrossRef] [PubMed]


| Phylum | Genus |
|---|---|
| Actinobacteria | Corynebacterium, Cutibacterium, Micrococcus |
| Firmicutes | Staphylococcus |
| Bacteroidetes | Flavobacteriales |
| Proteobacteria | Enhydrobacter, ß-Proteobacteria |
| Omics Technique | Definition | Application |
|---|---|---|
| (meta) Genomics/microbiomics | Culture-free analysis of the genetic heritage of an individual or a group of micro-organisms (meta) through innovative DNA extraction and replication techniques | Genotype detection and characterization of a microbial community |
| (meta) Transcriptomics | Sequencing of RNA (both mRNA and rRNA) present in a host’s cell or in a microbial community (meta) | Determination of the gene expression level and more in-depth knowledge of the active metabolic pathways |
| (meta) Proteomics | Protein abundances measurement and determination of protein species in the host’s cell or in the microbial community (meta) | Accurate analysis of the expression of functional active pathways |
| Metabolomics | Detection of host metabolites and by-products of microbial activity | Accurate snapshot of phenotypic expression and a deeper understanding of microbial communication |
| Authors/Years | Omics Technologies | Bio-Specimens | Technique | Results |
|---|---|---|---|---|
| Ashrafi et al. [52] 2020 | Metabolomic and metagenomic | Wound tissue | GC-MS 16sRNA amplicon | Temporal and dynamic acute wound metabolome and microbiome for identification of possible bio-markers that correspond to wound healing processes |
| Emmert et al. [53] 2020 | Lipidomics and metagenomic | Skin (by strip tape) | SFC-MS/MS 16sRNA amplicon | Cutaneous lipid composition alterations between body sites and correlations with the skin microbiome in AD, including in relation to FLG mutation |
| Kuehne et al. [54] 2017 | Metabolomic and transcriptomic | Epidermal tissue | QTOF−MS | Metabolic adaptations and its transcriptional regulation during human skin’s aging |
| Marathe et al. [55] 2021 | Metabolomic and proteomic | Epidermal tissue | LC-MS/MS 1H NMR | IL-9′s role as the main regulator of metabolic reprogramming and survival of KCs |
| Acharjee et al. [56] 2021 | Metabolomic, transcriptomic, and microRNAomic | Epidermal tissue Blood Serum | LC-MS targeted Microarray | Characteristic associations between genes, metabolites, and miRNAs in AD |
| Zhou et al. [57] 2017 | Metabolomic, proteomic and microRNAomic | Cultured Human keratinocyte HaCaT cells | UPLC/Q-TOF MS 2D-PAGE -MS Q-RT-PCR Microarray | Alterations in miRNA, protein, and metabolite profiles in arsenic-induced transformed cells, identifying potential early biomarkers for squamous cell carcinoma of the skin induced by arsenic exposure |
| Mirsa et al. [58] 2021 | Metabolomic, proteomic, and metagenomic | skin (by strip tape and sterile cotton-tipped dry swabs) and hair samples | UPLC-MS/MS HILIC/UPLC-MS/MS MRM/SRM GC–MS/MS LC–MS/MS 16sRNA and ITS1 amplicon | Macromolecular skin changes due to pollution could manifest as clinical signs of early skin pigmentation and/or other blemishes |
| Tilton et al. [59] 2015 | Metabolomic, transcriptomic, proteomic and phosphor-proteomic | In vitro 3D full-thickness human skin organotypic cultures | LC-MS GC-MS microarray | Identification of molecular responses and common pathways to low-dose radiation not highlighted by individual data sets, describing in detail the response mechanisms of complex biological systems |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dessì, A.; Pintus, R.; Fanos, V.; Bosco, A. Integrative Multiomics Approach to Skin: The Sinergy between Individualised Medicine and Futuristic Precision Skin Care? Metabolites 2024, 14, 157. https://doi.org/10.3390/metabo14030157
Dessì A, Pintus R, Fanos V, Bosco A. Integrative Multiomics Approach to Skin: The Sinergy between Individualised Medicine and Futuristic Precision Skin Care? Metabolites. 2024; 14(3):157. https://doi.org/10.3390/metabo14030157
Chicago/Turabian StyleDessì, Angelica, Roberta Pintus, Vassilios Fanos, and Alice Bosco. 2024. "Integrative Multiomics Approach to Skin: The Sinergy between Individualised Medicine and Futuristic Precision Skin Care?" Metabolites 14, no. 3: 157. https://doi.org/10.3390/metabo14030157
APA StyleDessì, A., Pintus, R., Fanos, V., & Bosco, A. (2024). Integrative Multiomics Approach to Skin: The Sinergy between Individualised Medicine and Futuristic Precision Skin Care? Metabolites, 14(3), 157. https://doi.org/10.3390/metabo14030157