Time-Course Metabolomic Analysis: Production of Betaine Structural Analogs by Fungal Fermentation of Seaweed
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Capillary Electrophoresis Mass Spectrometry (CE MS)
2.3. Statistical Analysis
3. Results
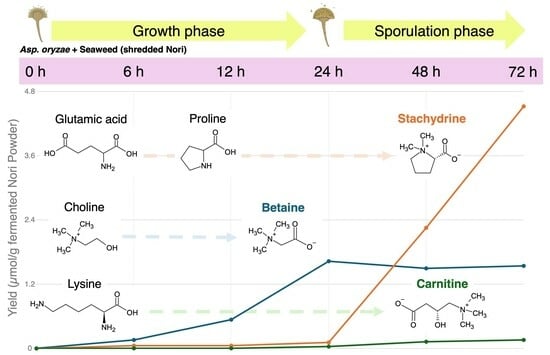
3.1. Metabolomic Changes in Nori Powder during Fungal Fermentation
3.2. Changes in Stachydrine Production and Related Metabolite Levels
3.3. Changes in Betaine Production and Related Metabolite Levels
3.4. Changes in Carnitine Production and Related Metabolite Levels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shang, X.; Pan, H.; Wang, X.; He, H.; Li, M. Leonurus japonicus Houtt.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2014, 152, 14–32. [Google Scholar] [CrossRef]
- Zhang, R.H.; Liu, Z.K.; Yang, D.S.; Zhang, X.J.; Sun, H.D.; Xiao, W.L. Phytochemistry and pharmacology of the genus Leonurus: The herb to benefit the mothers and more. Phytochemistry 2018, 147, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Zhou, Y.; Wang, M.; Guo, C.; Cao, Z.; Zhang, R.; Peng, C. A review of pharmacological and pharmacokinetic properties of stachydrine. Pharmacol. Res. 2020, 155, 104755. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Tang, Y.; Li, B.; Tang, J.; Xu, H.; Zhao, K.; Zhang, X. Stachydrine, a potential drug for the treatment of cardiovascular system and central nervous system diseases. Biomed. Pharmacother. 2023, 161, 114489. [Google Scholar] [CrossRef] [PubMed]
- Day, C.R.; Kempson, S.A. Betaine chemistry, roles, and potential use in liver disease. Biochim. Biophys. Acta 2016, 1860, 1098–1106. [Google Scholar] [CrossRef]
- Wang, C.; Ma, C.; Gong, L.; Dai, S.; Li, Y. Preventive and therapeutic role of betaine in liver disease: A review on molecular mechanisms. Eur. J. Pharmacol. 2021, 912, 174604. [Google Scholar] [CrossRef]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 370622. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.K.; Paal, M.C.; Donohue, T.M., Jr.; Ganesan, M.; Osna, N.A.; Kharbanda, K.K. Beneficial Effects of Betaine: A Comprehensive Review. Biology 2021, 10, 456. [Google Scholar] [CrossRef] [PubMed]
- Hanai, T.; Shiraki, M.; Imai, K.; Suetugu, A.; Takai, K.; Shimizu, M. Usefulness of Carnitine Supplementation for the Complications of Liver Cirrhosis. Nutrients 2020, 12, 1915. [Google Scholar] [CrossRef]
- Kepka, A.; Ochocinska, A.; Borzym-Kluczyk, M.; Skorupa, E.; Stasiewicz-Jarocka, B.; Chojnowska, S.; Waszkiewicz, N. Preventive Role of L-Carnitine and Balanced Diet in Alzheimer’s Disease. Nutrients 2020, 12, 1987. [Google Scholar] [CrossRef]
- Savic, D.; Hodson, L.; Neubauer, S.; Pavlides, M. The Importance of the Fatty Acid Transporter L-Carnitine in Non-Alcoholic Fatty Liver Disease (NAFLD). Nutrients 2020, 12, 2178. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhao, H. Role of Carnitine in Non-alcoholic Fatty Liver Disease and Other Related Diseases: An Update. Front. Med. 2021, 8, 689042. [Google Scholar] [CrossRef] [PubMed]
- Jahn, L.J.; Rekdal, V.M.; Sommer, M.O.A. Microbial foods for improving human and planetary health. Cell 2023, 186, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, K.I.; Yamagata, Y.; Tazawa, R.; Kitagawa, M.; Kato, T.; Isobe, K.; Kashiwagi, Y. Japanese Traditional Miso and Koji Making. J. Fungi 2021, 7, 579. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Matsuyama, A. Koji Molds for Japanese Soy Sauce Brewing: Characteristics and Key Enzymes. J. Fungi 2021, 7, 658. [Google Scholar] [CrossRef] [PubMed]
- Akaike, M.; Miyagawa, H.; Kimura, Y.; Terasaki, M.; Kusaba, Y.; Kitagaki, H.; Nishida, H. Chemical and Bacterial Components in Sake and Sake Production Process. Curr. Microbiol. 2020, 77, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Tamez-Hidalgo, P.; Cieplak, T.; Satessa, G.D.; Kot, W.; Kjærulff, S.; Nielsen, M.O.; Nielsen, D.S.; Krych, L. Supplementation of a lacto-fermented rapeseed-seaweed blend promotes gut microbial- and gut immune-modulation in weaner piglets. J. Anim. Sci. Biotechnol. 2021, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, S.R.; Oh, J.W. Effects of dietary fermented seaweed and seaweed fusiforme on growth performance, carcass parameters and immunoglobulin concentration in broiler chicks. Asian-Australas. J. Anim. Sci. 2014, 27, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Hideshima, N.; Araki, T. Development of koji by culturing Aspergillus oryzae on nori (Pyropia yezoensis). J. Biosci. Bioeng. 2019, 127, 183–189. [Google Scholar] [CrossRef]
- Nagao, K.; Inoue, N.; Tsuge, K.; Oikawa, A.; Kayashima, T.; Yanagita, T. Dried and Fermented Powders of Edible Algae (Neopyropia yezoensis) Attenuate Hepatic Steatosis in Obese Mice. Molecules 2022, 27, 2640. [Google Scholar] [CrossRef]
- Oikawa, A.; Matsuda, F.; Kikuyama, M.; Mimura, T.; Saito, K. Metabolomics of a single vacuole reveals metabolic dynamism in an alga Chara australis. Plant Physiol. 2011, 157, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xia, X.; Yuan, G.; Zhang, T.; Deng, B.; Feng, X.; Wang, Q. Stachydrine, a Bioactive Equilibrist for Synephrine, Identified from Four Citrus Chinese Herbs. Molecules 2023, 28, 3813. [Google Scholar] [CrossRef]
- Leete, E.; Marion, L.; Spenser, I.D. The biogenesis of alkaloids. XIII. The role of ornithine in the biosynthesis of stachydrine. J. Biol. Chem. 1955, 214, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Yun, J.H.; Lee, E.; Hong, S.P. Untargeted Metabolomics reveals Doenjang metabolites affected by manufacturing process and microorganisms. Food Res. Int. 2022, 157, 111422. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Kang, S.; Lee, A.R.; Kim, J.H.; Kim, T.W.; Lee, J.E.; Kim, H.R. Stachydrine derived from fermented rice prevents diet-induced obesity by regulating adipsin and endoplasmic reticulum homeostasis. J. Nutr. Biochem. 2022, 107, 109036. [Google Scholar] [CrossRef]
- Song, S.H.; Lee, C.; Lee, S.; Park, J.M.; Lee, H.J.; Bai, D.H.; Yoon, S.S.; Choi, J.B.; Park, Y.S. Analysis of microflora profile in Korean traditional nuruk. J. Microbiol. Biotechnol. 2013, 23, 40–46. [Google Scholar] [CrossRef]
- Strijbis, K.; Vaz, F.M.; Distel, B. Enzymology of the carnitine biosynthesis pathway. IUBMB Life 2010, 62, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Rousta, N.; Ferreira, J.A.; Taherzadeh, M.J. Production of L-carnitine-enriched edible filamentous fungal biomass through submerged cultivation. Bioengineered 2021, 12, 358–368. [Google Scholar] [CrossRef]
- Hamidi-Zad, Z.; Moslehi, A.; Rastegarpanah, M. Attenuating effects of allantoin on oxidative stress in a mouse model of nonalcoholic steatohepatitis: Role of SIRT1/Nrf2 pathway. Res. Pharm. Sci. 2021, 16, 651–659. [Google Scholar] [CrossRef]
- Apparoo, Y.; Phan, C.W.; Kuppusamy, U.R.; Sabaratnam, V. Ergothioneine and its prospects as an anti-ageing compound. Exp. Gerontol. 2022, 170, 111982. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Dai, X.; Cao, Y.; Wang, X.; Lu, J.; Xie, L.; Liu, K.; Li, X. The neuroprotective and antidiabetic effects of trigonelline: A review of signaling pathways and molecular mechanisms. Biochimie 2023, 206, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Kajiwara, Y.; Futagami, T.; Goto, M.; Takashita, H. Making Traditional Japanese Distilled Liquor, Shochu and Awamori, and the Contribution of White and Black Koji Fungi. J. Fungi 2021, 7, 517. [Google Scholar] [CrossRef] [PubMed]






| Metabolite | 6 h | 12 h | 24 h | 48 h | 72 h | Metabolite | 6 h | 12 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Betaine | 65.80 | 219.75 | 665.76 | 611.34 | 630.32 | 4-Methyl-5-thiazole-ethanol | 1.26 | 0.78 | 1.06 | 1.11 | 0.93 |
| Stachydrine | Appear | 1.02 | 2.12 | 42.58 | 85.38 | N-Acetylserine | 0.82 | 0.86 | 0.78 | 0.92 | 0.92 |
| Carnitine | Appear | 1.50 | 12.50 | 43.10 | 53.36 | Urate | 0.86 | 0.82 | 0.82 | 1.01 | 0.92 |
| Quinate | 1.20 | 1.01 | 2.22 | 18.19 | 41.84 | N-Acetyl-β-alanine | 0.95 | 0.93 | 0.81 | 1.20 | 0.87 |
| Allantoin | 1.05 | 1.11 | 5.52 | 30.87 | 37.76 | 5-Amino-4-oxovalerate | 0.95 | 0.93 | 0.81 | 1.20 | 0.87 |
| Uridine diphosphate-N-acetylgalactosamine | 1.07 | 1.47 | 5.67 | 18.04 | 33.04 | Serine | 0.93 | 0.97 | 0.78 | 0.66 | 0.83 |
| Uridine diphosphate-N-acetylglucosamine | 1.07 | 1.47 | 5.67 | 18.04 | 33.04 | Fructose-alanine | 0.99 | 1.22 | 1.00 | 1.09 | 0.81 |
| Nicotinate | 7.07 | 9.85 | 12.78 | 18.60 | 21.18 | 2-Isopropylmalate | 1.26 | 1.15 | 0.87 | 1.91 | 0.80 |
| Cytidine monophosphate_2 | n.d. | n.d. | Appear | 4.91 | 18.18 | 4-Methyl-2-oxovalerate | 1.26 | 1.15 | 0.87 | 1.91 | 0.80 |
| 3-Isopropylmalate | n.d. | n.d. | Appear | 8.48 | 17.75 | 4-Oxohexanoate | 1.26 | 1.15 | 0.87 | 1.91 | 0.80 |
| 5-Methylcytosine | n.d. | n.d. | Appear | 7.39 | 13.73 | 5-Oxohexanoate | 1.26 | 1.15 | 0.87 | 1.91 | 0.80 |
| S-Sulfocysteine | n.d. | n.d. | n.d. | Appear | 12.54 | Adenine | 0.61 | 0.44 | 0.67 | 0.44 | 0.75 |
| Thiamine | 2.62 | 5.89 | 13.97 | 13.23 | 10.18 | Diethanolamine_2 | 1.40 | 0.84 | 0.81 | 0.61 | 0.75 |
| Noradrenaline | n.d. | n.d. | Appear | 4.06 | 9.72 | Taurine | 0.87 | 0.81 | 0.81 | 0.94 | 0.72 |
| Xanthine | 2.72 | 6.15 | 8.00 | 19.53 | 9.43 | N6-Acetyllysine | 0.92 | 1.03 | 1.09 | 0.19 | 0.70 |
| Glucosamine-6-phosphate | n.d. | n.d. | Appear | 1.54 | 9.21 | Isocitrate | n.d. | n.d. | n.d. | Appear | 0.70 |
| Adenosine monophosphate_2 | 0.39 | 0.45 | 0.79 | 3.28 | 9.06 | Glucosamine | n.d. | n.d. | n.d. | Appear | 0.70 |
| N-Benzyloxycarbonylglycine | 1.14 | 0.97 | 1.54 | 5.01 | 8.59 | Galactosamine | n.d. | n.d. | n.d. | Appear | 0.70 |
| Glutamine | 1.27 | 1.28 | 5.23 | 0.62 | 8.59 | Lysine | 0.81 | 0.54 | 0.42 | 0.50 | 0.58 |
| Guanosine | 3.21 | 3.47 | 2.86 | 13.98 | 8.14 | Proline | 0.91 | 0.64 | 0.49 | 0.57 | 0.57 |
| Saccharopine | n.d. | Appear | 4.73 | 5.82 | 8.08 | Cytidine monophosphate_1 | 0.69 | 0.53 | 0.28 | 0.17 | 0.55 |
| Cysteine-glutathione disulfide | Disappear | n.d. | 2.20 | 3.19 | 7.98 | Galactose-1-phosphate | 1.20 | 0.88 | 0.27 | 0.41 | 0.54 |
| Deoxyguanosine | 0.81 | 1.12 | 1.00 | 5.97 | 7.58 | Glucose-1-phosphate | 1.20 | 0.88 | 0.27 | 0.41 | 0.54 |
| Hippurate | n.d. | n.d. | Appear | 6.78 | 7.38 | Inositol-1-phosphate | 1.20 | 0.88 | 0.27 | 0.41 | 0.54 |
| Trimethylamine N-Oxide | 0.80 | 1.13 | 0.78 | 2.83 | 7.33 | Mannose-1-phosphate | 1.20 | 0.88 | 0.27 | 0.41 | 0.54 |
| N-acetylgalactosamine | 1.39 | 1.10 | 1.68 | 5.08 | 6.18 | Citrate | 0.93 | 0.65 | 0.51 | 1.11 | 0.48 |
| N-acetylglucosamine | 1.39 | 1.10 | 1.68 | 5.08 | 6.18 | Succinate-semialdehyde | 0.76 | 0.66 | 0.58 | 0.75 | 0.47 |
| Phenylacetylglycine | 1.02 | 1.18 | 1.08 | 2.87 | 5.98 | Tyrosine | 0.91 | 0.90 | 0.47 | 0.38 | 0.47 |
| Raffinose | 2.38 | 2.92 | 3.04 | 5.51 | 5.89 | 5-Aminopentanoate | 0.91 | 0.97 | 1.00 | 0.86 | 0.39 |
| Cysteate | 1.02 | 0.78 | 1.19 | 4.87 | 5.68 | Threonine | 0.89 | 0.90 | 0.50 | 0.28 | 0.39 |
| Imidazole-4-acetate | n.d. | Appear | 1.22 | 3.12 | 4.94 | 2-Aminobutyrate | 0.96 | 0.88 | 0.38 | 0.26 | 0.37 |
| Hypoxanthine | 5.52 | 12.27 | 7.31 | 10.50 | 4.80 | 2-Aminoisobutyrate | 0.96 | 0.88 | 0.38 | 0.26 | 0.37 |
| 2-Aminoadipate | Appear | 2.14 | 4.09 | 3.17 | 4.51 | Glycine | 0.97 | 1.01 | 0.74 | 0.45 | 0.37 |
| N-acetylglucosamine-1-phosphate | 0.80 | 0.86 | 0.56 | 2.56 | 4.39 | Methylthioadenosine | Disappear | n.d. | n.d. | n.d. | 0.37 |
| Glutarate | 0.94 | 0.86 | 0.92 | 3.74 | 3.95 | Choline | 1.13 | 1.19 | 0.36 | 0.30 | 0.36 |
| Prostaglandin E2 | 0.96 | 2.44 | 17.94 | 6.27 | 3.86 | Adenosine | 0.11 | 0.07 | 0.18 | 0.18 | 0.34 |
| Sorbitol-6-phosphate | 1.34 | 0.94 | 0.54 | 2.17 | 3.18 | α-Ketoglutarate | 1.47 | 1.01 | 1.20 | 2.60 | 0.33 |
| 6-Phospho-gluconate | 1.79 | 1.17 | 3.05 | 4.51 | 3.15 | Tryptophan | 0.88 | 0.79 | 0.40 | 0.29 | 0.33 |
| Deoxycytidine 5′-diphosphate | 0.60 | 0.44 | 0.53 | 0.87 | 3.10 | 2,6-Diaminopimelate | 1.08 | 0.61 | 0.39 | 0.46 | 0.30 |
| Guanine | 1.55 | 2.55 | 1.58 | 3.06 | 3.06 | Arginine | 0.93 | 0.67 | 0.20 | 0.11 | 0.30 |
| Cytidine | 1.15 | 0.99 | 0.97 | 1.55 | 2.74 | Sedoheptulose-7-phosphate | 1.84 | 1.44 | 0.45 | 0.32 | 0.29 |
| N-acetylglucosamine-6-phosphate | Appear | 1.42 | 2.68 | 5.01 | 2.71 | Glutamic acid | 0.85 | 0.88 | 0.49 | 0.17 | 0.29 |
| N-acetylgalactosamine-6-phosphate | Appear | 1.42 | 2.68 | 5.01 | 2.71 | Phenylalanine | 0.95 | 0.83 | 0.35 | 0.28 | 0.29 |
| Calmodulin-like protein | n.d. | n.d. | Appear | 1.95 | 2.64 | N-Acetylornithine | 0.90 | 0.77 | 0.55 | 0.25 | 0.28 |
| Laurate | 1.00 | 0.48 | 8.19 | 7.90 | 2.39 | Pantothenate | 1.22 | 1.01 | 0.91 | 0.87 | 0.26 |
| 3-Aminoisobutyrate | Disappear | n.d. | 3.88 | 2.28 | 2.36 | Leucine | 0.96 | 0.82 | 0.35 | 0.29 | 0.26 |
| 3-Aminobutyrate | Disappear | n.d. | 3.88 | 2.28 | 2.36 | Aspartic acid | 0.75 | 0.75 | 0.49 | 0.13 | 0.26 |
| Methionine | 1.16 | 0.83 | 2.11 | 2.67 | 2.13 | Valine | 0.95 | 0.91 | 0.43 | 0.31 | 0.24 |
| Cytosine | 1.64 | 1.93 | 1.11 | 1.21 | 2.12 | Glycyl-L-leucine | 0.99 | 0.81 | 0.41 | 0.26 | 0.24 |
| 2-Hydroxybutyrate | 0.85 | 0.68 | 1.13 | 1.92 | 1.89 | Mannose-6-phosphate | 1.34 | 0.82 | 0.16 | 0.25 | 0.23 |
| 2-Hydroxyisobutyrate | 0.85 | 0.68 | 1.13 | 1.92 | 1.89 | Glucose-6-phosphate | 1.34 | 0.82 | 0.16 | 0.25 | 0.23 |
| Allantoate | 0.96 | 0.87 | 1.02 | 1.90 | 1.84 | Fructose-6-phosphate | 1.34 | 0.82 | 0.16 | 0.25 | 0.23 |
| Gluconate | 1.31 | 1.38 | 2.79 | 7.15 | 1.77 | Nicotinamide | 0.35 | 0.14 | 0.18 | 0.17 | 0.23 |
| Uridine diphosphate-galactose | 0.31 | 0.33 | 0.57 | 1.20 | 1.77 | Isonicotinamide | 0.35 | 0.14 | 0.18 | 0.17 | 0.23 |
| Uridine diphosphate-glucose | 0.31 | 0.33 | 0.57 | 1.20 | 1.77 | Trigonelline | n.d. | n.d. | Appear | 0.70 | 0.22 |
| Fructose-glutamic acid | 0.98 | 1.12 | 1.41 | 1.58 | 1.73 | Isoleucine | 0.95 | 0.86 | 0.34 | 0.25 | 0.22 |
| Cystathionine | n.d. | n.d. | Appear | 0.84 | 1.70 | Pyruvate | 3.04 | 2.64 | 0.48 | 0.86 | 0.21 |
| Decanoate | 1.32 | 0.78 | 2.96 | 3.10 | 1.67 | Glutamyl-L-alanine | 0.86 | 0.83 | 0.75 | 0.25 | 0.21 |
| Indole-3-acetic acid | n.d. | n.d. | n.d. | Appear | 1.64 | Pipecolate | 0.96 | 1.04 | 0.33 | 0.28 | 0.19 |
| Histidinol | 0.99 | 1.36 | 5.11 | 3.48 | 1.63 | N-Acetylglutamic acid | 1.00 | 0.82 | 0.66 | 0.50 | 0.18 |
| Nicotinamide adenine dinucleotide | n.d. | n.d. | 0.71 | 1.15 | 1.60 | Lactate | 0.76 | 0.52 | 0.22 | 0.24 | 0.18 |
| Galacturonate | 1.23 | 1.17 | 1.27 | 1.61 | 1.60 | Asparagine | 0.75 | 0.64 | 0.09 | 0.02 | 0.18 |
| Glucuronate | 1.23 | 1.17 | 1.27 | 1.61 | 1.60 | Glycerate | 0.92 | 0.83 | 0.61 | 0.28 | 0.17 |
| Histidine | 0.82 | 0.69 | 0.79 | 1.46 | 1.54 | Homoserine | 1.00 | 0.97 | 0.41 | 0.11 | 0.13 |
| Ergothioneine | 0.85 | 0.89 | 0.88 | 1.03 | 1.50 | Succinate | 1.29 | 0.82 | 0.37 | 0.62 | 0.13 |
| Mevalonolactone | 1.14 | 1.31 | 1.19 | 2.12 | 1.38 | Inosine monophosphate | 1.32 | 0.84 | 0.41 | 0.14 | 0.13 |
| Mevalonate | 1.14 | 1.31 | 1.19 | 2.12 | 1.38 | Adenosine diphosphate | 0.14 | 0.14 | 0.19 | n.d. | 0.12 |
| Pyridoxamine 5-phosphate | 1.13 | 1.24 | 0.65 | 0.48 | 1.32 | Guanosine monophosphate | 0.95 | 0.68 | 0.39 | 0.16 | 0.11 |
| Gibberellic acid | 1.25 | 0.92 | 0.96 | 1.03 | 1.31 | 4-Oxovalerate | 1.37 | 1.28 | 0.71 | 0.95 | 0.10 |
| 3-Hydroxybutyrate | 1.01 | 0.97 | 2.45 | 3.31 | 1.22 | Alanylalanine | 0.74 | 0.98 | 0.48 | 0.12 | 0.09 |
| Ornithine | 1.86 | 2.71 | 1.69 | 1.17 | 1.22 | 5-Oxoproline | 0.90 | 0.86 | 0.20 | 0.26 | 0.07 |
| Isethionate | 1.02 | 0.79 | 0.78 | 1.19 | 1.21 | Adenosine monophosphate_1 | 0.01 | 0.01 | 0.03 | 0.03 | 0.05 |
| Cystine | n.d. | n.d. | n.d. | Appear | 1.17 | Uridine monophosphate | 0.81 | 0.52 | 0.23 | 0.09 | 0.05 |
| Phenylpropanoate | 1.12 | 1.13 | 1.08 | 1.10 | 1.11 | Citrulline | 0.78 | 0.74 | 0.32 | 0.08 | 0.03 |
| 1-Methyladenosine | 0.85 | 0.78 | 0.52 | 0.68 | 1.04 | β-Alanine | 0.92 | 0.93 | 0.47 | 0.05 | 0.03 |
| Glycerol-3-phosphate | 1.21 | 1.16 | 0.89 | 1.46 | 1.03 | γ-Aminobutyric acid | 0.94 | 0.97 | 0.56 | 0.14 | 0.02 |
| Pyridoxal | n.d. | n.d. | Appear | 1.93 | 1.01 | Homoglutamine | 0.72 | 0.42 | 0.09 | 0.02 | 0.02 |
| Fumarate | 1.64 | 1.17 | 1.05 | 1.40 | 1.00 | Alanine | 0.93 | 0.89 | 0.22 | 0.04 | 0.02 |
| Sulfanilic acid | 1.08 | 0.95 | 0.94 | 1.36 | 1.00 | Phenethylamine | 0.92 | 0.68 | 0.39 | 0.39 | Disappear |
| Xanthosine monophosphate | n.d. | Appear | 0.87 | n.d. | n.d. | Ophthalmate | 4.18 | 3.97 | 0.38 | Disappear | n.d. |
| N-Acetylaspartic acid | n.d. | n.d. | n.d. | Appear | n.d. | Taurolithocholate | 0.79 | 0.42 | 0.38 | Disappear | n.d. |
| Guanidinosuccinate | n.d. | n.d. | n.d. | n.d. | Appear | Diethanolamine_1 | 0.91 | 0.99 | Disappear | n.d. | n.d. |
| N8-Acetylspermidine | n.d. | n.d. | n.d. | n.d. | Appear | N-Formylmethionine | 0.81 | 0.63 | Disappear | n.d. | n.d. |
| Ribulose-5-phosphate | n.d. | n.d. | n.d. | n.d. | Appear | Cytidine diphosphate | 0.33 | 0.22 | Disappear | n.d. | n.d. |
| Ribose-5-phosphate | n.d. | n.d. | n.d. | n.d. | Appear | Guanosine diphosphate-mannose | 0.22 | Disappear | n.d. | n.d. | n.d. |
| Xylulose-5-phosphate | n.d. | n.d. | n.d. | n.d. | Appear | Uridine diphosphate | 0.11 | Disappear | n.d. | n.d. | n.d. |
| N-Acetylmuramate | 1.44 | 1.15 | 0.86 | 1.20 | 0.99 | Guanosine diphosphate-glucose | 0.22 | Disappear | n.d. | n.d. | n.d. |
| Malate | 1.06 | 0.78 | 0.93 | 1.50 | 0.96 | S-adenosylhomocysteine | Disappear | n.d. | n.d. | n.d. | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inoue, N.; Tsuge, K.; Yanagita, T.; Oikawa, A.; Nagao, K. Time-Course Metabolomic Analysis: Production of Betaine Structural Analogs by Fungal Fermentation of Seaweed. Metabolites 2024, 14, 201. https://doi.org/10.3390/metabo14040201
Inoue N, Tsuge K, Yanagita T, Oikawa A, Nagao K. Time-Course Metabolomic Analysis: Production of Betaine Structural Analogs by Fungal Fermentation of Seaweed. Metabolites. 2024; 14(4):201. https://doi.org/10.3390/metabo14040201
Chicago/Turabian StyleInoue, Nao, Keisuke Tsuge, Teruyoshi Yanagita, Akira Oikawa, and Koji Nagao. 2024. "Time-Course Metabolomic Analysis: Production of Betaine Structural Analogs by Fungal Fermentation of Seaweed" Metabolites 14, no. 4: 201. https://doi.org/10.3390/metabo14040201
APA StyleInoue, N., Tsuge, K., Yanagita, T., Oikawa, A., & Nagao, K. (2024). Time-Course Metabolomic Analysis: Production of Betaine Structural Analogs by Fungal Fermentation of Seaweed. Metabolites, 14(4), 201. https://doi.org/10.3390/metabo14040201