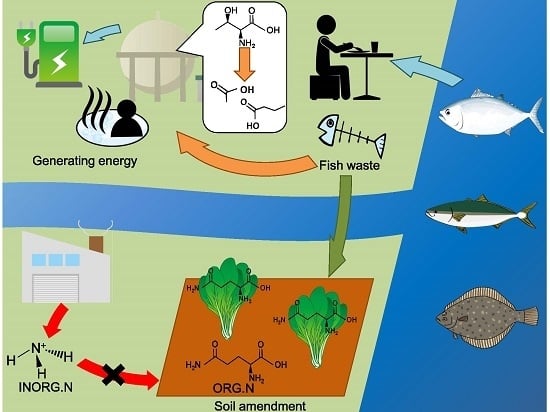
Visualization of Microfloral Metabolism for Marine Waste Recycling
Abstract
:1. Introduction

2. Results and Discussion
2.1. Characterization of Fish Waste

2.2. Metabolic Dynamics of Fish Waste by Microfloral Degradation

2.3. Amendment of Abandoned Agricultural Soils with Plant Growth

3. Experimental Section
3.1. Sample Preparation
3.2. Microfloral Degradation Experiments by Inputting with Fish Waste
3.3. Soil Amendment and Plant Growth Using Abandoned Agricultural Soils in the Tohoku Area
3.4. NMR Measurements
3.5. Elemental Analysis by ICP-OES (Inductively Coupled Plasma-Optical Emission Spectrometry) Measurement
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McCauley, D.J.; Pinsky, M.L.; Palumbi, S.R.; Estes, J.A.; Joyce, F.H.; Warner, R.R. Marine defaunation: Animal loss in the global ocean. Science 2015, 347, 248. [Google Scholar] [CrossRef] [PubMed]
- Oki, T.; Kanae, S. Global hydrological cycles and world water resources. Science 2006, 313, 1068–1072. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, X. Sustainability: Don’t waste seafood waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Rulli, M.C.; Saviori, A.; D’Odorico, P. Global land and water grabbing. Proc. Natl. Acad. Sci. USA 2013, 110, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Nakazoe, J.-I. Production and use of marine algae in japan. Jpn. Agric. Res. Q. 2001, 35, 281–290. [Google Scholar]
- López-Mosquera, M.E.; Fernández-Lema, E.; Villares, R.; Corral, R.; Alonso, B.; Blanco, C. Composting fish waste and seaweed to produce a fertilizer for use in organic agriculture. Procedia Environ. Sci. 2011, 9, 113–117. [Google Scholar] [CrossRef]
- Kramer, S.B.; Reganold, J.P.; Glover, J.D.; Bohannan, B.J.; Mooney, H.A. Reduced nitrate leaching and enhanced denitrifier activity and efficiency in organically fertilized soils. Proc. Natl. Acad. Sci. USA 2006, 103, 4522–4527. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, X. Sustainability: Don't waste seafood waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Gunaseelan, V.N. Anaerobic digestion of biomass for methane production: A review. Biomass Bioenergy 1997, 13, 83–114. [Google Scholar]
- Wang, J.L.; Wan, W. Factors influencing fermentative hydrogen production: A review. Int. J. Hydrog. Energy 2009, 34, 799–811. [Google Scholar] [CrossRef]
- Guo, X.M.; Trably, E.; Latrille, E.; Carrere, H.; Steyer, J.P. Hydrogen production from agricultural waste by dark fermentation: A review. Int. J. Hydrog. Energy 2010, 35, 10660–10673. [Google Scholar] [CrossRef]
- Date, Y.; Iikura, T.; Yamazawa, A.; Moriya, S.; Kikuchi, J. Metabolic sequences of anaerobic fermentation on glucose-based feeding substrates based on correlation analyses of microbial and metabolite profiling. J. Proteome Res. 2012, 11, 5602–5610. [Google Scholar] [CrossRef] [PubMed]
- Yamazawa, A.; Iikura, T.; Shino, A.; Date, Y.; Kikuchi, J. Solid-, solution-, and gas-state nmr monitoring of (1)(3)c-cellulose degradation in an anaerobic microbial ecosystem. Molecules 2013, 18, 9021–9033. [Google Scholar] [CrossRef] [PubMed]
- Yamazawa, A.; Iikura, T.; Morioka, Y.; Shino, A.; Ogata, Y.; Date, Y.; Kikuchi, J. Cellulose digestion and metabolism induced biocatalytic transitions in anaerobic microbial ecosystems. Metabolites 2013, 4, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Asano, R.; Nakai, Y.; Kawada, W.; Shimura, Y.; Inamoto, T.; Fukushima, J. Seawater inundation from the 2011 tohoku tsunami continues to strongly affect soil bacterial communities 1 year later. Microb. Ecol. 2013, 66, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Paustian, K.; Parton, W.J.; Persson, J. Modeling soil organic matter in organic-amended and nitrogen-fertilized long-term plots. Soil Sci. Soc. Am. J. 1992, 56, 476–488. [Google Scholar] [CrossRef]
- Rembialkowska, E. Organic Farming As a System to Provide Better Vegetable Quality. In Proceedings of the International Conference on Quality in Chains. An Integrated View on Fruit and Vegetable Quality, Wageningen, The Netherlands, 6–9 July 2003; pp. 473–479.
- Mader, P.; Fliessbach, A.; Dubois, D.; Gunst, L.; Fried, P.; Niggli, U. Soil fertility and biodiversity in organic farming. Science 2002, 296, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Hu, L.L.; Tang, J.J.; Wu, X.; Li, N.N.; Yuan, Y.G.; Yang, H.S.; Zhang, J.E.; Luo, S.M.; Chen, X. Ecological mechanisms underlying the sustainability of the agricultural heritage rice-fish coculture system. Proc. Natl. Acad. Sci. USA 2011, 108, E1381–E1387. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Date, Y.; Kikuchi, J. Differences in cellulosic supramolecular structure of compositionally similar rice straw affect biomass metabolism by paddy soil microbiota. PLoS ONE 2013, 8, e66919. [Google Scholar]
- Ogura, T.; Date, Y.; Tsuboi, Y.; Kikuchi, J. Metabolic dynamics analysis by massive data integration: Application to tsunami-affected field soils in japan. ACS Chem. Biol. 2015, 10, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Date, Y.; Akama, M.; Kikuchi, J. Comparative metabolomic and ionomic approach for abundant fishes in estuarine environments of japan. Sci. Rep. 2014, 4, 7005. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Sakata, K.; Yoshida, S.; Date, Y.; Kikuchi, J. Noninvasive analysis of metabolic changes following nutrient input into diverse fish species, as investigated by metabolic and microbial profiling approaches. PeerJ 2014, 2, e550. [Google Scholar] [CrossRef] [PubMed]
- Cloarec, O.; Dumas, M.E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1h nmr data sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.K.; Bong, Y.S.; Lee, K.S.; Hwang, G.S. An integrated analysis for determining the geographical origin of medicinal herbs using icp-aes/icp-ms and (1)h nmr analysis. Food Chem. 2014, 161, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Ha, M.; Lee, J.; Ahn, Y.G.; Kwak, J.H.; Ryu, D.H.; Hwang, G.S. Metabolite profiling of the response of burdock roots to copper stress. J. Agric. Food Chem. 2015, 63, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.K.; Bae, H.W.; Shin, S.K.; Jeon, T.W.; Seo, J.; Hwang, G.S. 1h-nmr-based profiling of organic components in leachate from animal carcasses disposal site with time. Environ. Sci. Pollut. Res. Int. 2014, 21, 10453–10460. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; An, Y.J.; Xu, W.J.; Kang, K.W.; Park, S. Real-time monitoring of cancer cell metabolism and effects of an anticancer agent using 2d in-cell nmr spectroscopy. Angew. Chem. Int. Ed. Engl. 2015, 54, 5374–5377. [Google Scholar] [CrossRef] [PubMed]
- Schlipalius, D.I.; Valmas, N.; Tuck, A.G.; Jagadeesan, R.; Ma, L.; Kaur, R.; Goldinger, A.; Anderson, C.; Kuang, J.; Zuryn, S.; et al. A core metabolic enzyme mediates resistance to phosphine gas. Science 2012, 338, 807–810. [Google Scholar] [CrossRef] [PubMed]
- An, Y.J.; Xu, W.J.; Jin, X.; Wen, H.; Kim, H.; Lee, J.; Park, S. Metabotyping of the c. Elegans sir-2.1 mutant using in vivo labeling and (13)c-heteronuclear multidimensional nmr metabolomics. ACS Chem. Biol. 2012, 7, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.; Lane, A.N. Nmr-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR 2011, 49, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.; Tan, J.; McKinney, M.M.; Lane, A.N. Stable isotope resolved metabolomics analysis of ribonucleotide and rna metabolism in human lung cancer cells. Metabolomics 2012, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Lee, T.; You, S.; Park, S.H.; Song, H.; Eilber, K.S.; Anger, J.T.; Freeman, M.R.; Park, S.; Kim, J. Urinary metabolite profiling combined with computational analysis predicts interstitial cystitis-associated candidate biomarkers. J. Proteome Res. 2015, 14, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Shino, A.; Akashi, K.; Kikuchi, J. Chemical profiling of jatropha tissues under different torrefaction conditions: Application to biomass waste recovery. PLoS ONE 2014, 9, e106893. [Google Scholar]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 25 January 2016).
- Chikayama, E.; Suto, M.; Nishihara, T.; Shinozaki, K.; Kikuchi, J. Systematic nmr analysis of stable isotope labeled metabolite mixtures in plant and animal systems: Coarse grained views of metabolic pathways. PLoS ONE 2008, 3, e3805. [Google Scholar] [CrossRef] [PubMed]
- Chikayama, E.; Sekiyama, Y.; Okamoto, M.; Nakanishi, Y.; Tsuboi, Y.; Akiyama, K.; Saito, K.; Shinozaki, K.; Kikuchi, J. Statistical indices for simultaneous large-scale metabolite detections for a single nmr spectrum. Anal. Chem. 2010, 82, 1653–1658. [Google Scholar] [CrossRef] [PubMed]
- Date, Y.; Nakanishi, Y.; Fukuda, S.; Nuijima, Y.; Kato, T.; Umehara, M.; Ohno, H.; Kikuchi, J. In vitro evaluation method for screening of candidate prebiotic foods. Food Chem. 2014, 152, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Misawa, T.; Date, Y.; Kikuchi, J. Human metabolic, mineral, and microbiota fluctuations across daily nutritional intake visualized by a data-driven approach. J. Proteome Res. 2015, 14, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.M.; Moriya, S.; Tsuboi, Y.; Date, Y.; Prieto-da-Silva, A.R.; Radis-Baptista, G.; Yamane, T.; Kikuchi, J. Biogeochemical typing of paddy field by a data-driven approach revealing sub-systems within a complex environment—A pipeline to filtrate, organize and frame massive dataset from multi-omics analyses. PLoS ONE 2014, 9, e110723. [Google Scholar]
- Asakura, T.; Date, Y.; Kikuchi, J. Comparative analysis of chemical and microbial profiles in estuarine sediments sampled from kanto and tohoku regions in japan. Anal. Chem. 2014, 86, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Sakata, K.; Date, Y.; Kikuchi, J. Integrated analysis of seaweed components during seasonal fluctuation by data mining across heterogeneous chemical measurements with network visualization. Anal. Chem. 2014, 86, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Ito, K.; Sakata, K.; Date, Y.; Kikuchi, J. Pretreatment and integrated analysis of spectral data reveal seaweed similarities based on chemical diversity. Anal. Chem. 2015, 87, 2819–2826. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogura, T.; Hoshino, R.; Date, Y.; Kikuchi, J. Visualization of Microfloral Metabolism for Marine Waste Recycling. Metabolites 2016, 6, 7. https://doi.org/10.3390/metabo6010007
Ogura T, Hoshino R, Date Y, Kikuchi J. Visualization of Microfloral Metabolism for Marine Waste Recycling. Metabolites. 2016; 6(1):7. https://doi.org/10.3390/metabo6010007
Chicago/Turabian StyleOgura, Tatsuki, Reona Hoshino, Yasuhiro Date, and Jun Kikuchi. 2016. "Visualization of Microfloral Metabolism for Marine Waste Recycling" Metabolites 6, no. 1: 7. https://doi.org/10.3390/metabo6010007
APA StyleOgura, T., Hoshino, R., Date, Y., & Kikuchi, J. (2016). Visualization of Microfloral Metabolism for Marine Waste Recycling. Metabolites, 6(1), 7. https://doi.org/10.3390/metabo6010007