Unravelling the Metabolic Reconfiguration of the Post-Challenge Primed State in Sorghum bicolor Responding to Colletotrichum sublineolum Infection
Abstract
:1. Introduction
2. Results and Discussion
2.1. Evaluation of Anthracnose Symptom Development in P. alvei Primed vs. Naïve Sorghum Plants Challenged with the Hemibiotrophic Pathogen, Colletotrichum sublineolum
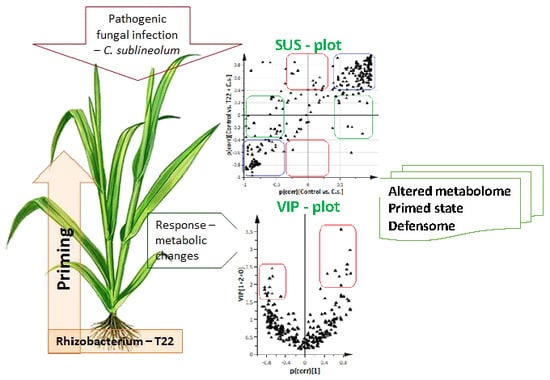
2.2. Comparative Analysis of the Metabolomic Profiles of P. alvei (T22)-Primed and Naïve Sorghum Plants Challenged with the Anthracnose Pathogen, Colletotrichum sublineolum
2.3. Differential Defence-Related Metabolic Changes in P. alvei Primed vs. Naïve Sorghum Plants Challenged with the Hemibiotrophic Pathogen, Colletotrichum sublineolum
2.3.1. Differential Changes in Primary Metabolism and Plant Hormones Levels
2.3.2. Defence Responses in Colletotrichum sublineolum-Challenged (Primed vs. Naïve) Sorghum Plants: Differential Changes in the Lipidome and Phenolics
3. Materials and Methods
3.1. Preparation of Sorghum Seedlings and Colletotrichum sublineolum Spore Suspensions
3.2. Plant Growth Promoting Rhizobacteria Preparation and Inoculation of the Sorghum Seedlings
3.3. Secondary Challenge: Inoculation of Sorghum Seedlings with C. Sublineolum
3.4. Metabolite Extraction and Analyses by Ultrahigh Performance Liquid Chromatography-High Definition-Mass Spectrometry (UHPLC-HD-MS)
3.5. Data Analysis: Data Set Matrix Creation and Chemometric Analyses
3.6. Metabolite Annotation: Putative Identification of Chemometrically Selected Metabolites
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Mhlongo, M.; Piater, L.; Dubery, I. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [PubMed]
- van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef]
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Beneloujaephajri, E.; Costa, A.; Haridon, F.L.; Métraux, J.; Binda, M. Production of reactive oxygen species and wound-induced resistance in Arabidopsis thaliana against Botrytis cinerea are preceded and depend on a burst of calcium. BMC Plant Biol. 2013, 13, 160. [Google Scholar] [CrossRef]
- Cohen, Y.; Vaknin, M.; Mauch-Mani, B. BABA-induced resistance: Milestones along a 55-year journey. Phytoparasitica 2016, 44, 513–538. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Sánchez-Bel, P.; Flors, V. Molecular and physiological stages of priming: How plants prepare for environmental challenges. Plant. Cell Rep. 2014, 33, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Mareya, C.R.; Tugizimana, F.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defence-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019, 9, 1840. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Labuschagne, N. Differential metabolic reprogramming in Paenibacillus alvei-primed Sorghum bicolor seedlings in response to Fusarium pseudograminearum infection. Metabolites 2019, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Piater, L.A.; Dubery, I.A. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 18–20. [Google Scholar] [CrossRef]
- Beisken, S.; Eiden, M.; Salek, R.M. Getting the right answers: Understanding metabolomics challenges. Expert Rev. Mol. Diagn. 2015, 15, 97–109. [Google Scholar] [CrossRef]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Labuschagne, N.; Pretorius, T.; Hassen, A.I. Plant Growth Promoting Rhizobacteria as Biocontrol Agents Against Soil-Borne Plant Diseases. In Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18, pp. 211–230. ISBN 978-3-642-13611-5. [Google Scholar]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Ji, H.; Liu, X.S. Analyzing’ omics data using hierarchical models. Nat. Biotechnol. 2010, 28, 337–340. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Grissa, D.; Pétéra, M.; Brandolini, M.; Napoli, A.; Comte, B.; Pujos-Guillot, E. Feature Selection methods for early predictive biomarker discovery using untargeted metabolomic data. Front. Mol. Biosci. 2016, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Steenkamp, P.; Piater, L.; Dubery, I. A Conversation on data mining strategies in lc-ms untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Daghir-Wojtkowiak, E.; Kaliszan, R.; Markuszewski, M.J. PLS-based and regularization-based methods for the selection of relevant variables in non-targeted metabolomics data. Front. Mol. Biosci. 2016, 3, 35. [Google Scholar] [CrossRef]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chemom. 2014, 28, 8. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Wheelock, A.M.; Wheelock, C.E. Trials and tribulations of “omics” data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef]
- Wilkinson, L. Dot Plots. Am. Stat. 1999, 53, 276–281. [Google Scholar] [CrossRef]
- Pastor, V.; Balmer, A.; Gamir, J.; Flors, V.; Mauch-Mani, B. Preparing to fight back: Generation and storage of priming compounds. Front. Plant Sci. 2014, 5, 295. [Google Scholar] [CrossRef]
- Chen, Y.H.; Bernal, C.C.; Tan, J.; Horgan, F.G.; Fitzgerald, M.A. Planthopper “adaptation” to resistant rice varieties: Changes in amino acid composition over time. J. Insect Physiol. 2011, 57, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. Lignification: Flexibility, biosynthesis and regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vallet, A.; Ramos, B.; Bednarek, P.; Lopez, G.; Pislewska-Bednarek, M.; Schulze-Lefert, P.; Molina, A. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 2010, 63, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, A.; Hashimoto, Y.; Tanaka, C.; Dubouzet, J.G.; Nakao, T.; Matsuda, F.; Nishioka, T.; Miyagawa, H.; Wakasa, K. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008, 54, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Iven, T.; Ko, S.; Singh, S.; Braus-stromeyer, S.A.; Bischoff, M.; Tietze, L.F.; Braus, G.H.; Lipka, V.; Feussner, I.; Droge-Laser, W. Transcriptional activation and production of tryptophan-derived secondary metabolites in Arabidopsis roots contributes to the defense against the fungal vascular pathogen Verticillium longisporum. Mol. Plant 2012, 5, 1389–1402. [Google Scholar] [CrossRef]
- Finnegan, T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. The lipopolysaccharide-induced metabolome signature in Arabidopsis thaliana reveals dynamic reprogramming of phytoalexin and phytoanticipin Pathways. PLoS ONE 2016, 11, e0163572. [Google Scholar] [CrossRef]
- Newman, M.A.; von Roepenack-Lahaye, E.; Parr, A.; Daniels, M.J.; Dow, J.M. Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol. Plant Microbe Interact. 2001, 14, 785–792. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Celenza, J.L. Metabolism of tyrosine and tryptophan—New genes for old pathways. Curr. Opin. Plant Biol. 2001, 4, 234–240. [Google Scholar] [CrossRef]
- Busk, P.K.; Møller, B.L. Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol. 2002, 129, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Glauser, G. Family business: Multiple members of major phytohormone classes orchestrate plant stress responses. Chem.-A Eur. J. 2010, 16, 10280–10289. [Google Scholar] [CrossRef] [PubMed]
- Kohli, A.; Sreenivasulu, N.; Lakshmanan, P.; Kumar, P.P. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013, 32, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 2016, 56, 174–189. [Google Scholar] [CrossRef] [PubMed]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Ahn, I.-P.; Lee, S.-W.; Suh, S.-C. Rhizobacteria-induced priming in Arabidopsis is dependent on ethylene, jasmonic acid, and NPR1. Mol. Plant Microbe. Interact. 2007, 20, 759–768. [Google Scholar] [CrossRef]
- Gozzo, F.; Faoro, F. Systemic acquired resistance (50 years after discovery): Moving from the lab to the field. J. Agric. Food Chem. 2013, 61, 12473–12491. [Google Scholar] [CrossRef] [PubMed]
- Brien, J.A.O.; Benková, E. Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 2013, 4, 451. [Google Scholar] [CrossRef] [Green Version]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Knirsch, V.; Körber, N.; Pieruschka, R.; Fiorani, F.; Brzobohatý, B.; Černý, M.; Spichal, L.; et al. Cytokinins: Their impact on molecular and growth responses to drought stress and recovery in Arabidopsis. Front. Plant Sci. 2018, 9, 655. [Google Scholar] [CrossRef] [PubMed]
- Gajdosova, S.; Spichal, L.; Kaminek, M.; Hoyerova, K.; Novak, O.; Dobrev, P.I.; Galuszka, P.; Klima, P.; Gaudinova, A.; Zizkova, E.; et al. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, M.; Brütting, C.; Meza-canales, I.D.; Großkinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef]
- Reusche, M.; Klásková, J.; Thole, K.; Truskina, J.; Janz, D.; Strnad, M.; Spíchal, L.; Lipka, V.; Teichmann, T. Stabilization of cytokinin levels enhances Arabidopsis resistance against Verticillium longisporum. Mol. Plant-Microbe Interact. 2013, 26, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Veselova, S.V.; Farkhutdinov, R.G.; Veselov, D.S.; Kudoyarova, G.R. Role of cytokinins in the regulation of stomatal conductance of wheat seedlings under conditions of rapidly changing local temperature. Russ. J. Plant Physiol. 2006, 54, 756–761. [Google Scholar] [CrossRef]
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Gan, P.; Ikeda, K.; Irieda, H.; Narusaka, M.; O’Connell, R.J.; Narusaka, Y.; Takano, Y.; Kubo, Y.; Shirasu, K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 2013, 197, 1236–1249. [Google Scholar] [CrossRef]
- Kubo, Y.; Harata, K.; Kodama, S.; Fukada, F. Development of the infection strategy of the hemibiotrophic plant pathogen, Colletotrichum orbiculare, and plant immunity. Physiol. Mol. Plant Pathol. 2016, 95, 32–36. [Google Scholar] [CrossRef]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Fahrmann, J.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Time-resolved decoding of metabolic signatures of in vitro growth of the hemibiotrophic pathogen Colletotrichum sublineolum. Sci. Rep. 2019, 9, 3290. [Google Scholar] [CrossRef] [PubMed]
- Berkey, R.; Bendigeri, D.; Xiao, S. Sphingolipids and plant defense/disease: The “death” connection and beyond. Front. Plant Sci. 2012, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Siebers, M.; Brands, M.; Wewer, V.; Duan, Y.; Hölzl, G.; Dörmann, P. Lipids in plant–microbe interactions. Biochim. Biophys. Acta 2016, 1861, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Robin, G.P. Systemic signaling during plant defense. Curr. Opin. Plant Biol. 2013, 16, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Pata, M.O.; Hannun, Y.A.; Ng, C.K.-Y. Plant sphingolipids: Decoding the enigma of the Sphinx. New Phytol. 2010, 185, 611–630. [Google Scholar] [CrossRef]
- Michaelson, L.V.; Napier, J.A.; Molino, D.; Faure, J. Plant sphingolipids: Their importance in cellular organization and adaption. Biochim. Biophys. Acta 2016, 1861, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Peer, M.; Stegmann, M.; Mueller, M.J.; Waller, F. Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010, 584, 4053–4056. [Google Scholar] [CrossRef]
- Coursol, S.; Le Stunff, H.; Lynch, D.V.; Gilroy, S.; Assmann, S.M.; Spiegel, S. Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 2005, 137, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Magnin-Robert, M.; Le Bourse, D.; Markham, J.; Dorey, S.; Clément, C.; Baillieul, F.; Dhondt-Cordelier, S. Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in Arabidopsis. Plant Physiol. 2015, 169, 2255–2274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef]
- Snyder, B.A.; Leite, B.; Hipskind, J.; Butler, L.G.; Nicholson, R.L. Accumulation of sorghum phytoalexins induced by Colletotrichum graminicola at the infection site. Physiol. Mol. Plant Pathol. 1991, 39, 463–470. [Google Scholar] [CrossRef]
- Poloni, A.; Schirawski, J. Red card for pathogens: Phytoalexins in sorghum and maize. Molecules 2014, 19, 9114–9133. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef] [PubMed]
- Hassen, A.I.; Labuschagne, N.; Korsten, L. Efficacy of rhizobacteria for growth promotion in sorghum under greenhouse conditions and selected modes of action studies. J. Agric. Sci. 2009, 147, 17–30. [Google Scholar] [CrossRef]
- Madala, N.E.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Collision energy alteration during mass spectrometric acquisition is essential to ensure unbiased metabolomics-based analyses. Anal. Bioanal. Chem. 2012, 404, 367–372. [Google Scholar] [CrossRef]
- Nelson, P.R.C.; Taylor, P.A.; MacGregor, J.F. Missing data methods in PCA and PLS: Score calculations with incomplete observations. Chemom. Intell. Lab. Syst. 1996, 35, 45–65. [Google Scholar] [CrossRef]
- Bro, R.; Kjeldahl, K.; Smilde, A.K.; Kiers, H.A.L. Cross-validation of component models: A critical look at current methods. Anal. Bioanal. Chem. 2008, 390, 1241–1251. [Google Scholar] [CrossRef]
- Brown, M.; Wedge, D.C.; Goodacre, R.; Kell, D.B.; Baker, P.N.; Kenny, L.C.; Mamas, M.A.; Neyses, L.; Dunn, W.B. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics 2011, 27, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Dunn, W.B.; Dobson, P.; Patel, Y.; Winder, C.L.; Francis-McIntyre, S.; Begley, P.; Carroll, K.; Broadhurst, D.; Tseng, A.; et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst 2009, 134, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Price, W.E.; Ashton, J.; Tapsell, L.C.; Johnson, S. Identification and characterization of phenolic compounds in hydromethanolic extracts of sorghum wholegrains by LC-ESI-MSn. Food Chem. 2016, 211, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Metabolites | m/z | Rt (min) | Adduct | ESI Mode | Molecular Formula | Biochemical Classification | Post-Challenge Period (Primed/Naïve) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–3 d.p.i. | 5–9 d.p.i. | ||||||||||
| p-Value | Fold Change | p-Value | Fold Change | ||||||||
| 1 | L-Tyrosine | 182.0819 | 1.25 | H | pos | C9H11NO3 | Amino acid | 7.97 × 10−7 | 88.41 | 1.68 × 10−10 | 0.52 |
| 2 | 5-Hydroxytryptophan | 236.1036 | 2.65 | NH3 | neg | C11H12N2O3 | Amino acid | 1.33 × 10−10 | 89.28 | 1.34 × 10−11 | 1.99 |
| 3 | L-Tryptophan | 205.0978 | 3.02 | H | pos | C11H12N2O2 | Amino acid | 5.91 × 10−11 | 2.85 | 2.48 × 10−10 | 0.57 |
| 4 | Dhurrin | 329.1335 | 4.02 | NH3 | pos | C14H17NO7 | Cyanogenic glucoside | 7.68 × 10−27 | 40.50 | 4.91 × 10−8 | 24.80 |
| 5 | Naringin chalcone | 627.1912 | 2.52 | HCOOH | neg | C27H34O14 | Flavonoid | 1.50 × 10−7 | 6.13 | 3.34 × 10−10 | 1.30 |
| 6 | Naringin | 625.1761 | 3.46 | HCOOH | neg | C27H32O14 | Flavonoid | 5.17 × 10−7 | 5.46 | 3.60 × 10−10 | 2.08 |
| 7 | Peptahydroxychalcone 4′-O-glucoside | 449.1067 | 4.73 | H | neg | C21H22O11 | Flavonoid | 1.88 × 10−15 | 2.29 | 8.14 × 10−10 | 1.60 |
| 8 | Hesperidin | 609.1809 | 5.00 | H | neg | C28H34O15 | Flavonoid | 3.43 × 10−11 | 2.22 | 2.36 × 10−9 | 0.74 |
| 9 | Apigenin 7-O-[beta-d-apiosyl-(1->2)-beta-d-glucoside] | 563.1390 | 5.05 | H | neg | C26H28O14 | Flavonoid | 4.51 × 10−4 | 1.09 | 1.03 × 10−14 | 4.84 |
| 10 | Kaempferol 3-O-rhamnoside-7-O-glucoside | 593.1501 | 5.68 | H | neg | C27H30O15 | Flavonoid | 2.75 × 10−4 | 3.24 | 1.53 × 10−14 | 0.36 |
| 11 | Cyanidin 3-O-rhamnosylglucoside | 595.1657 | 5.75 | H | pos | C27H30O15 | Flavonoid | 1.74 × 10−14 | 0.34 | 4.06 × 10−19 | 0.22 |
| 12 | Kaempferol-3-glucoside | 447.0921 | 5.88 | H | neg | C21H20O11 | Flavonoid | 1.97 × 10−4 | 3.22 | 2.84 × 10−14 | 4.94 |
| 13 | Quercitrin | 449.1080 | 5.92 | H | pos | C21H20O11 | Flavonoid | 2.08 × 10−4 | 1.14 | 2.28 × 10−14 | 0.22 |
| 14 | Apigenin | 271.1544 | 6.02 | H | pos | C15H10O5 | Flavonoid | 8.82 × 10−4 | 0.30 | 2.01 × 10−7 | 0.36 |
| 15 | Apigeninidin | 255.1533 | 6.10 | H | pos | C15H11O4 | Flavonoid | 6.84 × 10−24 | 0.31 | 1.10 × 10−32 | 0.77 |
| 16 | Luteolin 7-O-beta-d-glucoside | 447.0921 | 6.19 | H | neg | C21H20O11 | Flavonoid | 8.27 × 10−5 | 1.12 | 2.95 × 10−12 | 0.27 |
| 17 | Apigenin 7-O-neohesperidoside | 579.1709 | 6.27 | H | pos | C27H30O14 | Flavonoid | 1.86 × 10−11 | 1.00 | 3.06 × 10−31 | 0.20 |
| 18 | Luteolin | 287.0536 | 6.30 | H | pos | C15H10O6 | Flavonoid | 4.25 × 10−23 | 0.36 | 0.000 | 0.51 |
| 19 | 1,2-bis-O-Sinapoyl-beta-d-glucoside | 591.1705 | 6.35 | H | neg | C28H32O14 | Flavonoid | 1.28 × 10−29 | 3.17 | 5.02 × 10−14 | 10.39 |
| 20 | 7-O-Methylvitexin 2′′-O-beta-l-rhamnoside | 615.1680 | 6.39 | Na | pos | C28H32O14 | Flavonoid | 4.35 × 10−13 | 0.31 | 1.09 × 10−25 | 0.07 |
| 21 | Isovitexin 2′′-O-beta-d-glucoside | 593.1501 | 6.68 | H | neg | C27H30O15 | Flavonoid | 7.22 × 10−3 | 1.96 | 1.08 × 10−14 | 3.65 |
| 22 | Luteolinidin | 271.0616 | 6.87 | H | pos | C15H11O5 | Flavonoid | 8.23 × 10−3 | 0.36 | 7.68 × 10−27 | 0.68 |
| 23 | 12,13-Epoxy-9-hydroxy-10-octadecenoate | 395.2040 | 9.26 | HCOONa | neg | C18H32O5 | Lipid | 1.52 × 10−17 | 50.98 | 5.07 × 10−4 | 39.73 |
| 24 | Phytosphingosine | 318.3009 | 10.52 | H | pos | C18H39NO3 | Lipid | 1.29 × 10−29 | 267.28 | 0.000 | 27.50 |
| 25 | 16-Hydroxypalmitate | 290.2700 | 10.58 | NH3 | pos | C16H32O3 | Lipid | 7.30 × 10−12 | 217.16 | 1.42 × 10−19 | 24.76 |
| 26 | (9Z)-(13S)-12,13-Epoxyoctadeca-9,11-dienoic acid | 363.2137 | 11.44 | HCOONa | pos | C18H30O3 | Lipid | 1.96 × 10−14 | 139.22 | 8.66 × 10−11 | 35.53 |
| 27 | 13(S)-hydroxyperoxy-octadecatrienoic acid | 309.2071 | 11.79 | H | neg | C18H30O4 | Lipid | 1.02 × 10−10 | 40.98 | 1.84 × 10−5 | 28.80 |
| 28 | 25-Hydroxy-24-epi-brassinolide | 519.3267 | 13.34 | Na | pos | C28H48O7 | Lipid | 9.77 × 10−14 | 35.74 | 1.47 × 10−6 | 28.16 |
| 29 | Oleanolate 3-beta-d-glucuronoside-28-glucoside | 795.4497 | 15.36 | H | pos | C42H66O14 | Lipid | 2.32 × 10−22 | 31.31 | 1.94 × 10−14 | 19.49 |
| 30 | Oleanoic acid 3-O-glucuronide | 655.3820 | 15.40 | Na | pos | C36H56O9 | Lipid | 0.000 | 36.35 | 1.58 × 10−37 | 18.05 |
| 31 | Caffeoylquinate | 377.0846 | 3.83 | Na | pos | C16H18O9 | Phenylpropanoid | 3.64 × 10−12 | 12.59 | 1.35 × 10−28 | 9.05 |
| 32 | p-Coumaroyl quinic acid | 427.0621 | 1.03 | NaHCOONa | neg | C16H18O8 | Phenylpropanoid | 4.06 × 10−6 | 13.20 | 1.94 × 10−27 | 6.75 |
| 33 | Feruloyltyramine | 331.1650 | 2.01 | NH3 | pos | C18H19NO4 | Phenylpropanoid | 5.02 × 10−14 | 10.39 | 2.38 × 10−20 | 11.63 |
| 34 | 4-Coumaroylshikimate | 319.1062 | 3.16 | H | neg | C16H16O7 | Phenylpropanoid | 2.46 × 10−7 | 12.33 | 3.41 × 10−19 | 0.58 |
| 35 | 2-Coumarate | 165.0554 | 3.25 | H | pos | C9H8O3 | Phenylpropanoid | 2.16 × 10−17 | 8.78 | 2.20 × 10−20 | 0.58 |
| 36 | 1-O-Sinapoyl-beta-d-glucose | 387.1279 | 3.56 | H | pos | C17H22O10 | Phenylpropanoid | 3.23 × 10−9 | 14.50 | 6.72 × 10−20 | 0.59 |
| 37 | 4-O-beta-d-Glucosyl-4-hydroxycinnamate | 395.0947 | 4.09 | HCOONa | pos | C15H18O8 | Phenylpropanoid | 7.40 × 10−5 | 14.12 | 9.53 × 10−20 | 1.60 |
| 38 | Ferulate | 209.0448 | 4.58 | H | neg | C10H10O5 | Phenylpropanoid | 9.42 × 10−6 | 12.13 | 5.37 × 10−25 | 0.47 |
| 39 | O-Feruloylquinate | 367.1017 | 4.88 | H | neg | C17H20O9 | Phenylpropanoid | 2.38 × 10−20 | 11.63 | 9.62 × 10−23 | 0.49 |
| 40 | Coniferyl acetate | 291.0844 | 1.09 | HCOONa | pos | C12H14O4 | Phenylpropanoid | 9.28 × 10−7 | 12.52 | 9.86 × 10−19 | 2.41 |
| 41 | Zeatin | 220.1192 | 2.38 | H | pos | C10H13N5O | Phytohormone | 0.000 | 17.09 | 3.19 × 10−21 | 14.17 |
| 42 | Salicylate-glucoside | 299.0758 | 1.79 | H | neg | C13H16O8 | Phytohormone | 1.43 × 10−4 | 9.19 | 2.01 × 10−17 | 1.66 |
| 43 | 6-Hydroxy-indole-3-acetyl-phenylalanine | 405.1077 | 2.76 | HCOONa | neg | C19H18N2O4 | Phytohormone | 1.38 × 10−4 | 9.36 | 1.51 × 10−17 | 2.75 |
| 44 | 6-Hydroxy-indole-3-acetyl-valine | 335.0962 | 2.82 | Na_Na | pos | C15H18N2O4 | Phytohormone | 3.08 × 10−6 | 10.71 | 2.83 × 10−16 | 4.88 |
| 45 | (-)-Jasmonoyl-l-isoleucine | 406.1626 | 4.33 | HCOOK | neg | C18H29NO4 | Phytohormone | 6.08 × 10−9 | 8.21 | 2.16 × 10−17 | 8.78 |
| 46 | 12-Hydroxyjasmonic acid 12-O-beta-d-glucoside | 429.1514 | 5.59 | Na_Na | neg | C19H30O8 | Phytohormone | 1.57 × 10−6 | 10.73 | 1.14 × 10−16 | 2.76 |
| 47 | trans-Zeatin-7-beta-d-glucoside | 399.1990 | 8.14 | NH3 | pos | C16H23N5O6 | Phytohormone | 6.82 × 10−6 | 9.53 | 2.98 × 10−16 | 0.46 |
| 48 | Riboflavin | 419.0969 | 5.80 | Na_Na | neg | C17H20N4O6 | Riboflavin | 1.42 × 10−19 | 24.76 | 2.84 × 10−7 | 18.67 |
| 49 | Feruloylserotonin | 351.1333 | 11.66 | H | neg | C20H20N2O4 | Trp pathway | 1.44 × 10−3 | 23.73 | 1.09 × 10−25 | 15.32 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the Metabolic Reconfiguration of the Post-Challenge Primed State in Sorghum bicolor Responding to Colletotrichum sublineolum Infection. Metabolites 2019, 9, 194. https://doi.org/10.3390/metabo9100194
Tugizimana F, Steenkamp PA, Piater LA, Labuschagne N, Dubery IA. Unravelling the Metabolic Reconfiguration of the Post-Challenge Primed State in Sorghum bicolor Responding to Colletotrichum sublineolum Infection. Metabolites. 2019; 9(10):194. https://doi.org/10.3390/metabo9100194
Chicago/Turabian StyleTugizimana, Fidele, Paul A. Steenkamp, Lizelle A. Piater, Nico Labuschagne, and Ian A. Dubery. 2019. "Unravelling the Metabolic Reconfiguration of the Post-Challenge Primed State in Sorghum bicolor Responding to Colletotrichum sublineolum Infection" Metabolites 9, no. 10: 194. https://doi.org/10.3390/metabo9100194
APA StyleTugizimana, F., Steenkamp, P. A., Piater, L. A., Labuschagne, N., & Dubery, I. A. (2019). Unravelling the Metabolic Reconfiguration of the Post-Challenge Primed State in Sorghum bicolor Responding to Colletotrichum sublineolum Infection. Metabolites, 9(10), 194. https://doi.org/10.3390/metabo9100194