Deciphering the Resistance Mechanism of Tomato Plants Against Whitefly-Mediated Tomato Curly Stunt Virus Infection through Ultra-High-Performance Liquid Chromatography Coupled to Mass Spectrometry (UHPLC-MS)-Based Metabolomics Approaches
Abstract
:1. Introduction
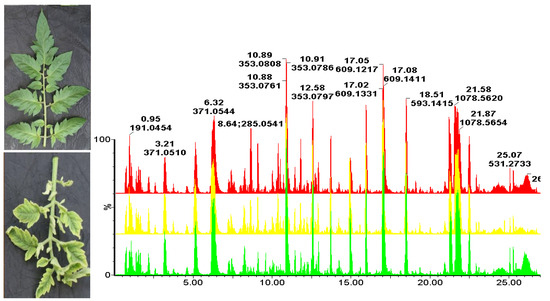
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Experimental Design and Plant Treatments
- Con (no treatment): a seedling tray, with 18 d old S and RT seedlings, received no whitefly or viral treatment and served as a control.
- WF (mock-inoculation): identical seedling tray of S and RT seedlings were placed into a cage. Whiteflies, which had been reared on healthy (i.e., virus-free) cotton and tomato plants (and had not been allowed to acquire the virus), were transferred onto the seedlings. These non-viruliferous whiteflies were allowed to feed on seedlings for four days.
- WF + Vir (whitefly-mediated ToCSV infection): in order to distinguish the metabolic response caused by viral infection and that caused by whitefly feeding alone, another identical seedling tray of 18 d old S and RT seedlings was placed into a different cage and the viruliferous whitefly transferred from a ToCSV infected cultivar (Rooikhaki) to the seedlings, where after the infected plant material was removed. The whiteflies were then allowed a four-day inoculation access period (IAP) on the seedlings. After four days, the whiteflies were removed from the seedlings, by shaking, and all the seedling trays, Con, WF, and WF + Vir moved to another room, where they were subjected to contact and systemic insecticide.
4.3. Plant Material Collection
4.4. Metabolite Extraction
4.5. UHPLC-ESI-MS Analysis
4.6. Multivariate Data Analysis
4.7. Metabolite Annotation and Semi-Quantitation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- FAOSTAT. Value of Agricultural Production. Available online: http://www.fao.org/faostat/en/#data/QV (accessed on 29 May 2017).
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16, 181–190. [Google Scholar] [CrossRef]
- Bauchet, G.; Causse, M. Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives. In Genetic Diversity in Plants; Caliskan, M., Ed.; InTechOpen: Shanghai, China, 2012; pp. 134–162. [Google Scholar]
- ICTV, International Committee on Taxonomy of Viruses. 2017. Available online: http://www.ictvonline.org/virusTaxonomy.asp (accessed on 13 November 2017).
- Polston, J.E.; Anderson, P.K. The emergence of whitefly-transmitted geminiviruses in tomato in the western hemisphere. Plant Dis. 1997, 81, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging viral diseases of tomato crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Leke, W.N.; Mignouna, D.B.; Brown, J.K.; Kvarnheden, A. Begomovirus disease complex: Emerging threat to vegetable production systems of West and Central Africa. Agric. Food Secur. 2015, 4, 1. [Google Scholar] [CrossRef]
- Gill, R.J.; Brown, J.K. Systematics of Bemisia and Bemisia relatives: Can molecular techniques solve the Bemisia tabaci complex conundrum-a taxonomist’s viewpoint. In Bemisia: Bionomics and Management of a Global Pest; Stansly, P.A., Naranjo, S.E., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 5–29. [Google Scholar]
- Tay, W.T.; Evans, G.A.; Boykin, L.M.; de Barro, P.J. Will the real Bemisia tabaci please stand up? PLoS ONE 2012, 7, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Perring, T.M. The Bemisia tabaci species complex. Crop Prot. 2001, 20, 725–737. [Google Scholar] [CrossRef]
- Boykin, L.M.; Shatters, R.G.; Rosell, R.C.; McKenzie, C.L.; Bagnall, R.A.; De Barro, P.; Frohlich, D.R. Global relationships of Bemisia tabaci (hemiptera: aleyrodidae) revealed using bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogenet. Evol. 2007, 44, 1306–1319. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Lapidot, M.; Friedmann, M. Breeding for resistance to whitefly-transmitted geminiviruses. Ann. Appl. Biol. 2002, 140, 109–127. [Google Scholar] [CrossRef]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Zakay, Y.; Navot, N.; Zeidan, M.; Kedar, N.; Rabinowitch, H.; Czosnek, H.; Zamir, D. Screening Lycopersicon accessions for resistance to Tomato yellow leaf curl virus: Presence of viral DNA and symptom development. Plant Dis. 1991, 72, 279–281. [Google Scholar] [CrossRef]
- Vidavsky, F.; Czosnek, H. Tomato breeding lines resistant and tolerant to Tomato yellow leaf curl virus issued from Lycopersicon hirsutum. Phytopathology 1998, 88, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.M.; Bernacchi, D.; Green, S.; Tanksley, S.D.; Muniyappa, V.; Attiganal, P.S.; Chen, H.; Kuo, G.; Fang, D.; Chen, J. Mapping a wild tomato introgression associated with Tomato yellow leaf curl virus resistance in a cultivated tomato line. J. Am. Soc. Hortic. Sci. 2000, 125, 15–20. [Google Scholar] [CrossRef]
- Schauer, N.; Zamir, D.; Fernie, A.R. Metabolic profiling of leaves and fruit of wild species tomato: A survey of the Solanum lycopersicum complex. J. Exp. Bot. 2005, 56, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Semel, Y.; Roessner, U.; Gur, A.; Balbo, I.; Carrari, F.; Pleban, T.; Perez-Melis, A.; Bruedigam, C.; Kopka, J.; et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006, 24, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Moshe, A.; Pfannstiel, J.; Brotman, Y.; Kolot, M.; Sobol, I.; Czosnek, H.; Gorovits, R. Stress responses to Tomato yellow leaf curl virus (TYLCV) infection of resistant and susceptible tomato plants are different. Metabolomics 2012. [Google Scholar] [CrossRef]
- Osbourn, A.E. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 1996, 8, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.E.; Qi, X.; Townsend, B.; Qin, B. Dissecting plant secondary metabolism - constitutive chemical defences in cereals. New Phytol. 2003, 159, 101–108. [Google Scholar] [CrossRef]
- Iriti, M.; Faoro, F. Chemical diversity and defence metabolism: How plants cope with pathogens and ozone pollution. Int. J. Mol. Sci. 2009, 10, 3371–3399. [Google Scholar] [CrossRef]
- Gust, A.A.; Brunner, F.; Nürnberger, T. Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 2010, 21, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.V.G.; Engel, B.; de Vos, R.C.H.; Vereijken, P.; Astola, L.; Groenenboom, M.; van de Geest, H.; Bovy, A.G.; Molenaar, J.; van Eeuwijk, F.A.; et al. Metabolomics reveals organ-specific metabolic rearrangements during early tomato seedling development. Metabolomics 2014, 10, 958–974. [Google Scholar] [CrossRef]
- Johnson, H.E.; Broadhurst, D.; Goodacre, R.; Smith, A.R. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry 2003, 62, 919–928. [Google Scholar] [CrossRef]
- Abu-Nada, Y.; Kushalappa, A.C.; Marshall, W.D.; Al-Mughrabi, K.; Murphy, A. Temporal dynamics of pathogenesis-related metabolites and their plausible pathways of induction in potato leaves following inoculation with Phytophthora infestans. Eur. J. Plant Pathol. 2007, 118, 375–391. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Faubert, D.; Jabaji, S.A. Metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE 2014, 9, e111930. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Vaclavikova, M.; Dzuman, Z.; Vaclavik, L.; Ovesna, J.; Hajslova, J. Rapid LC-MS-based metabolomics method to study the Fusarium infection of barley. J. Sep. Sci. 2014, 37, 912–919. [Google Scholar] [CrossRef]
- Pushpa, D.; Yogendra, K.N.; Gunnaiah, R.; Kushalappa, A.C.; Murphy, A. Identification of late blight resistance-related metabolites and genes in potato through nontargeted metabolomics. Plant Mol. Biol. Rep. 2014, 32, 584–595. [Google Scholar] [CrossRef]
- Eloh, K.; Sasanelli, N.; Maxia, A.; Caboni, P. Untargeted metabolomics of tomato plants after root-knot nematode infestation. J. Agric. Food Chem. 2016, 64, 5963–5968. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Lisón, P.; Kim, H.K.; Choi, Y.H.; Verpoorte, R.; Rodrigo, I.; Conejero, V.; Bellés, J.M. Metabolic fingerprinting of Tomato Mosaic Virus infected Solanum lycopersicum. J. Plant Physiol. 2012, 169, 1586–1596. [Google Scholar] [CrossRef]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- Bagherian, S.A.A.; Hamzehzarghani, H.; Izadpanah, K.; Djavaheri, M. Effects of potato spindle tuber viroid infection on tomato metabolic profile. J. Plant Physiol. 2016, 201, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, A.D.; Gómez, S.; Osorio, S.; Fernie, A.R.; Orians, C.M. Herbivore-induced changes in tomato (Solanum lycopersicum) primary metabolism: A whole plant perspective. J. Chem. Ecol. 2011, 37, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.R.; Ament, K.; Sabelis, M.W.; Haring, M.A.; Schuurink, R.C. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 2004, 135, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Errard, A.; Ulrichs, C.; Kuühne, S.; Mewis, I.; Drungowski, M.; Schreiner, M.; Baldermann, S. Single versus multiple-pest infestation affects differently the biochemistry of tomato (Solanum lycopersicum ‘Ailsa Craig’). J. Agric. Food Chem. 2015, 63, 10103–10111. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.; Rudaz, S.; Choi, Y.H.; Kim, H.K. Plant metabolomics: From holistic data to relevant biomarkers. Curr. Med. Chem. 2013, 20, 1056–1090. [Google Scholar] [PubMed]
- Pietersen, G.; Idris, A.M.; Krüger, K.; Brown, J.K. Characterization of Tomato curly stunt virus: A new tomato-infecting begomovirus from South Africa. Plant Pathol. 2008, 57, 809–818. [Google Scholar] [CrossRef]
- Hammerschmidt, R. Chlorogenic acid: A versatile defense compound. Physiol. Mol. Plant Pathol. 2014, 88, 3–8. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE 2016, 11, e0151350. [Google Scholar] [CrossRef]
- Bostock, R.M.; Wilcox, S.M.; Wang, G.; Adaskaveg, J.E. Suppression of Monilinia fructicola cutinase production by peach fruit surface phenolic acids. Physiol. Mol. Plant Pathol. 1999, 54, 37–50. [Google Scholar] [CrossRef]
- Terry, L.A.; Joyce, D.C.; Adikaram, N.K.B.; Khambay, B.P.S. Preformed antifungal compounds in strawberry fruit and flower tissues. Postharvest Biol. Technol. 2004, 31, 201–212. [Google Scholar] [CrossRef]
- Ruelas, C.; Tiznado-Hernandez, M.E.; Sanchez-Estrada, A.; Robles-Burgueno, M.R.; Troncoso-Rojas, R. Changes in phenolic acid content during Alternaria alternata infection in tomato fruit. J. Phytopathol. 2006, 154, 236–244. [Google Scholar] [CrossRef]
- Pietersen, G.; Smith, M. Tomato yellow leaf curl virus resistant tomatoes show resistance to Tomato curly stunt virus. Plant Dis. 2002, 86, 528–534. [Google Scholar] [CrossRef]
- Clifford, M.N.; Johnston, K.L.; Knight, S.; Kuhnert, N. Hierarchical scheme for LC-MSn identification of chlorogenic acids. J. Agric. Food Chem. 2003, 51, 2900–2911. [Google Scholar] [CrossRef]
- Clifford, M.N.; Knight, S.; Kuhnert, N. Discriminating between the six isomers of dicaffeoylquinic acid by LC-MSn. J. Agric. Food Chem. 2005, 53, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N.; Kirkpatrick, J.; Kuhnert, N.; Roozendaal, H.; Salgado, P.R. LC-MSn analysis of the cis isomers of chlorogenic acid. Food Chem. 2008, 106, 379–385. [Google Scholar] [CrossRef]
- Ncube, E.; Mhlongo, M.; Piater, L.; Steenkamp, P.; Dubery, I.A.; Madala, N.E. Analyses of chlorogenic acids and related cinnamic acid derivatives from Nicotiana tabacum tissues with the aid of UPLC-QTOF-MS/MS based on the in-source collision-induced dissociation method. Chem. Cent. J. 2014, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Dictionary of Natural Products (DNP). Available online: Dnp.chemnetbase.com (accessed on 31 May 2017).
- Camañes, G.; Scalschi, L.; Vicedo, B.; González-Bosch, C.; García-Agustínet, P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 2015, 84, 125–139. [Google Scholar] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical AnalysisWorking Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- XCMS-online Metabolomics. Available online: https://xcmsonline.scripps.edu/ (accessed on 31 January 2017).
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, P.; Steenkamp, P.A.; Dubery, I.A. Comparative metabolic phenotyping of tomato (Solanum lycopersicum) for the identification of metabolic signatures in cultivars differing in resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. [Google Scholar] [CrossRef]





| m/z [M−H]− | Rt (Min) | Elemental Formula | Annotation/Identity | Treatment, Line (RT vs. S) and Day (8, 15, 25, 35) that Metabolite Was Identified as a Biomarker | ||
|---|---|---|---|---|---|---|
| Con | WF | Wf + Vir | ||||
| 163.0395 | 10.80 | C9H8O3 | Coumaric acid | # | # | RT-8 |
| 191.0556 | 10.59 | C7H12O6 | Quinic acid | RT-8, S-8 | RT-8, S-8 | # |
| 209.0297 | 7.79 | C6H10O8 | Galactaric acid | RT-8 | RT-8, S-8 | # |
| 221.0189 | 16.01 | - | Unknown b | # | RT-8, S-8 | RT-8, S-8 |
| 245.0926 | 13.91 | C13H14N2O3 | Acetyl tryptophan | # | RT-8, S-8 | S-8 |
| 249.1150 | 6.93 | C13H18N2O3 | Caffeoyl putrecine | # | RT-8 | S-8 |
| 251.0867 | 17.13 | - | Unknown b | # | # | RT-8, S-8 |
| 254.0360 | 13.08 | - | Unknown b | RT-8 | # | RT-8, S-8 |
| 270.0311 | 11.77 | - | Unknown b | S-8 | # | RT-8, S-8 |
| 284.0532 | 8.25 | C12H13O8 | 2,3-Dihydroxybenzoic-3-O-β-D-xyloside a | # | RT-8, S-8 | RT-8, S-8, S-35 |
| 291.1261 | 8.34 | - | Unknown b | # | S-35 | # |
| 291.1251 | 10.28 | - | Unknown b | # | S-35 | S-35 |
| 307.1210 | 8.82 | - | Unknown b | # | S-35 | S-35 |
| 321.1361 | 9.84 | - | Unknown b | # | S-35 | S-35 |
| 351.1212 | 11.42 | - | Unknown b | # | # | RT-8, S-8, S-35 |
| 353.0873 | 7.89 | C16H18O9 | 3-O-Caffeoylquinic acid a | S-8 | S-8, S-35 | S-35 |
| 353.0873 | 10.58 | C16H18O9 | 5-O-(E)-Caffeoylquinic acid a | RT-8, S-8, S-35 | RT-8, S-8, S-35 | # |
| 353.0873 | 10.58 | C16H18O9 | 5-O-(E)-Caffeoylquinic acid a | # | RT-8, S-8 | # |
| 353.0873 | 11.09 | C16H18O9 | 4-O-Caffeoylquinic acid a | RT-8, S-8, S-35 | RT-8, S-8, S-35 | # |
| 353.0873 | 12.24 | C16H18O9 | 5-O-(Z)-Caffeoylquinic acid a | RT-8, S-8 | # | # |
| 355.1029 | 11.76 | C16H20O9 | Ferulic acid glycoside a | S-35 | # | S-35 |
| 365.0089 | 7.11 | - | Unknown b | # | RT-8, S-8 | RT-8, S-8, S-35 |
| 367.1029 | 13.41 | C17H20O9 | 5-O-Feruloylquinic acid a | RT-8, S-8, S-35 | S-8 | # |
| 371.0614 | 7.77 | C15H16O11 | 5-O-Caffeoylgalactaric acid a | RT-8, S-8, S-35 | RT-8, S-8, S-35 | # |
| 385.1135 | 13.30 | C17H22O10 | Sinapoylglycoside a | S-8, S-35 | RT-8, S-8, S-35 | # |
| 401.1448 | 12.49 | C18H26O10 | Benzyl alcohol-hexose-pentosea | # | RT-8, S-8 | RT-8, S-8, S-35 |
| 431.1849 | 13.30 | - | Sinapoylglycoside FA a | # | RT-8, S-8 | S-8 |
| 447.1440 | 12.57 | - | Benzyl alcohol-hexose-pentose FA a | # | # | S-35 |
| 571.1299 | 8.38 | C24H28O16 | 2,3-Dihydroxybenzoic-3-O-β-D-xyloside (dimer) a | # | # | S-35 |
| 583.2713 | 10.27 | - | Unknown (dimer of 291) b | # | S-35 | S-35 |
| 593.1472 | 18.10 | - | Unknown | RT-8, S-8, S-35 | # | # |
| 609.1456 | 16.66 | C27H30O16 | Quercetin 3-rutinoside a | RT-8, S-8, S-35 | S-35 | RT-8, S-8, S-35 |
| 615.2621 | 8.81 | - | Unknown (dimer of 307) b | # | S-35 | S-35 |
| 675.1130 | 6.99 | C22H30NO23 | b | # | RT-8, S-8 | RT-8 |
| 677.1507 | 19.49 | C34H30O15 | b | # | RT-8, S-8 | RT-8, S-8 |
| 693.3487 | 14.7 | C37H50N4O9 | N1,N4,N12 tris (dihydroxycaffeoyl)spermine | # | RT-8 | RT-8, S-8 |
| 707.1823 | 10.58 | C32H36O18 | 5-O-(E)-Caffeoylquinic acid (dimer) a | RT-8, S-8, S-35 | RT-8, S-8 | # |
| 735.2136 | 13.53 | C34H40O18 | 5-O-(E)-Feruloylquinic acid (dimer) a | # | # | S-35 |
| 741.1878 | 15.59 | C32H38O20 | Quercetin-3-O-deoxyhexose-O-hexose-O-pentose a | RT-8, S-8 | S-35 | # |
| 743.1307 | 7.78 | C15H16O11 | 5-O-(E)-Caffeoylgalactaric acid (dimer) a | RT-8, S-8, S-35 | RT-8, S-8 | # |
| 771.1984 | 12.76 | C33H40O21 | Quercetin-3-rutinoside-7-glycoside a | S-35 | # | # |
| 947.2514 | 18.41 | - | Unknown b | S-35 | S-35 | # |
| 1094.5383 | 19.09 | C51H85NO24 | Hydroxytomatine (Lycoperoside H)-FA | RT-8 | RT-8, S-8, S-35 | S-35 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossouw, L.T.; Madala, N.E.; Tugizimana, F.; Steenkamp, P.A.; Esterhuizen, L.L.; Dubery, I.A. Deciphering the Resistance Mechanism of Tomato Plants Against Whitefly-Mediated Tomato Curly Stunt Virus Infection through Ultra-High-Performance Liquid Chromatography Coupled to Mass Spectrometry (UHPLC-MS)-Based Metabolomics Approaches. Metabolites 2019, 9, 60. https://doi.org/10.3390/metabo9040060
Rossouw LT, Madala NE, Tugizimana F, Steenkamp PA, Esterhuizen LL, Dubery IA. Deciphering the Resistance Mechanism of Tomato Plants Against Whitefly-Mediated Tomato Curly Stunt Virus Infection through Ultra-High-Performance Liquid Chromatography Coupled to Mass Spectrometry (UHPLC-MS)-Based Metabolomics Approaches. Metabolites. 2019; 9(4):60. https://doi.org/10.3390/metabo9040060
Chicago/Turabian StyleRossouw, Leandri T., Ntakadzeni E. Madala, Fidele Tugizimana, Paul A. Steenkamp, Lindy L. Esterhuizen, and Ian A. Dubery. 2019. "Deciphering the Resistance Mechanism of Tomato Plants Against Whitefly-Mediated Tomato Curly Stunt Virus Infection through Ultra-High-Performance Liquid Chromatography Coupled to Mass Spectrometry (UHPLC-MS)-Based Metabolomics Approaches" Metabolites 9, no. 4: 60. https://doi.org/10.3390/metabo9040060
APA StyleRossouw, L. T., Madala, N. E., Tugizimana, F., Steenkamp, P. A., Esterhuizen, L. L., & Dubery, I. A. (2019). Deciphering the Resistance Mechanism of Tomato Plants Against Whitefly-Mediated Tomato Curly Stunt Virus Infection through Ultra-High-Performance Liquid Chromatography Coupled to Mass Spectrometry (UHPLC-MS)-Based Metabolomics Approaches. Metabolites, 9(4), 60. https://doi.org/10.3390/metabo9040060