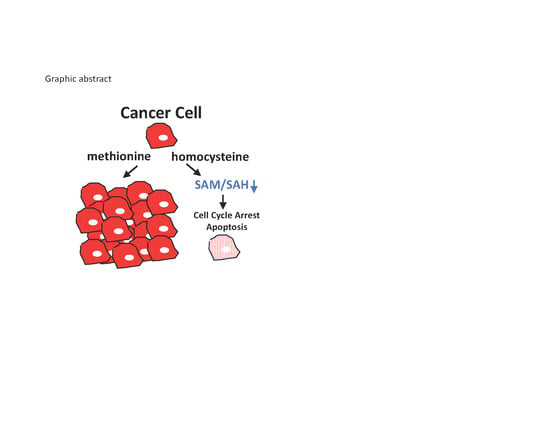
Methionine Dependence of Cancer
Abstract
:1. Introduction
2. Methionine Metabolism
3. Polyamine Synthesis and Methionine Metabolism
4. Methionine Metabolism and Cancer
5. A Metabolic Cell Cycle Checkpoint Related to Methionine Metabolism
6. The SAM Checkpoint in Yeast
7. The SAM Checkpoint in Mammals
8. Methionine Metabolism and Tumor Growth
9. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Sugimura, T.; Birnbaum, S.M.; Winitz, M.; Greenstein, J.P. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acide. Arch. Biochem. Biophys. 1959, 81, 448–455. [Google Scholar] [CrossRef]
- Chello, P.L.; Bertino, J.R. Dependence of 5-methyltetrahydrofolate utilization by L5178Y murine leukemia cells in vitro on the presence of hydroxycobalamin and transcobalamin II. Cancer Res. 1973, 33, 1898–1904. [Google Scholar] [PubMed]
- Hoffman, R.M.; Erbe, R.W. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl. Acad. Sci. USA 1976, 73, 1523–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halpern, B.C.; Clark, B.R.; Hardy, D.N.; Halpern, R.M.; Smith, R.A. The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc. Natl. Acad. Sci. USA 1974, 71, 1133–1136. [Google Scholar] [CrossRef] [Green Version]
- Stern, P.H.; Wallace, C.D.; Hoffman, R.M. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J. Cell. Physiol. 1984, 119, 29–34. [Google Scholar] [CrossRef]
- Mecham, J.O.; Rowitch, D.; Wallace, C.D.; Stern, P.H.; Hoffman, R.M. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem. Biophys. Res. Commun. 1983, 117, 429–434. [Google Scholar] [CrossRef]
- Booher, K.; Lin, D.W.; Borrego, S.L.; Kaiser, P. Downregulation of Cdc6 and pre-replication complexes in response to methionine stress in breast cancer cells. Cell Cycle 2012, 11, 4414–4423. [Google Scholar] [CrossRef] [Green Version]
- Borrego, S.L.; Fahrmann, J.; Datta, R.; Stringari, C.; Grapov, D.; Zeller, M.; Chen, Y.; Wang, P.; Baldi, P.; Gratton, E.; et al. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 2016, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.-W.; Chung, B.P.; Kaiser, P. S-adenosylmethionine limitation induces p38 mitogen-activated protein kinase and triggers cell cycle arrest in G1. J. Cell Sci. 2014, 127, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Lien, E.C.; Ghisolfi, L.; Geck, R.C.; Asara, J.M.; Toker, A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 2017, 10, pii:eaao6604. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.-W.; Kaiser, P.; University of California, Irvine. Unpublished work. 2020.
- Su, Y.-C.; Kaiser, P.; University of California, Irvine. Unpublished work. 2020.
- Lu, S.; Epner, D.E. Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr. Cancer 2000, 38, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Kotb, M.; Kredich, N.M. Regulation of human lymphocyte S-adenosylmethionine synthetase by product inhibition. Biochim. Biophys. Acta 1990, 1039, 253–260. [Google Scholar] [CrossRef]
- Halim, A.-B.B.; LeGros, L.; Geller, A.; Kotb, M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J. Biol. Chem. 1999, 274, 29720–29725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeGros, H.L.; Geller, A.M.; Kotb, M. Differential Regulation of Methionine Adenosyltransferase in Superantigen and Mitogen Stimulated Human T Lymphocytes. J. Biol. Chem. 1997, 272, 16040–16047. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: http://www.proteinatlas.org (accessed on 8 March 2020).
- Coward, J.K.; Slisz, E.P. Analogs of S-adenosylhomocysteine as potential inhibitors of biological transmethylation. Specificity of the S-adenosylhomocysteine binding site. J. Med. Chem. 1973, 16, 460–463. [Google Scholar] [CrossRef]
- Loenen, W.A. S-adenosylmethionine: Jack of all trades and master of everything? Biochem. Soc. Trans. 2006, 34, 330–333. [Google Scholar] [CrossRef] [Green Version]
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Pegg, A.E.; Casero, R.A., Jr. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar]
- Soda, K. Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism. Int. J. Mol. Sci. 2018, 19, 3106. [Google Scholar] [CrossRef] [Green Version]
- Kryukov, G.V.; Wilson, F.H.; Ruth, J.R.; Paulk, J.; Tsherniak, A.; Marlow, S.E.; Vazquez, F.; Weir, B.A.; Fitzgerald, M.E.; Tanaka, M.; et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 2016, 351, 1214–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lattouf, H.; Poulard, C.; Le Romancer, M. PRMT5 prognostic value in cancer. Oncotarget 2019, 10, 3151–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.H.; Zhang, H.; Savarese, T.M. Gene deletion chemoselectivity: Codeletion of the genes for p16(INK4), methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy. Cancer Res. 1996, 56, 1083–1090. [Google Scholar] [PubMed]
- Karikari, C.A.; Mullendore, M.; Eshleman, J.R.; Argani, P.; Leoni, L.M.; Chattopadhyay, S.; Hidalgo, M.; Maitra, A. Homozygous deletions of methylthioadenosine phosphorylase in human biliary tract cancers. Mol. Cancer Ther. 2005, 4, 1860–1866. [Google Scholar] [CrossRef] [Green Version]
- Nobori, T.; Takabayashi, K.; Tran, P.; Orvis, L.; Batova, A.; Yu, A.L.; Carson, D.A. Genomic cloning of methylthioadenosine phosphorylase: A purine metabolic enzyme deficient in multiple different cancers. Proc. Natl. Acad. Sci. USA 1996, 93, 6203–6208. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Batova, A.; Shao, L.; Carrera, C.J.; Yu, A.L. Presence of methylthioadenosine phosphorylase (MTAP) in hematopoietic stem/progenitor cells: Its therapeutic implication for MTAP (-) malignancies. Clin. Cancer Res. 1997, 3, 433–438. [Google Scholar]
- Hoffman, R.M.; Jacobsen, S.J.; Erbe, R.W. Reversion to methionine independence by malignant rat and SV40-transformed human fibroblasts. Biochem. Biophys. Res. Commun. 1978, 82, 228–234. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Jacobsen, S.J.; Erbe, R.W. Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc. Natl. Acad. Sci. USA 1979, 76, 1313–1317. [Google Scholar] [CrossRef] [Green Version]
- Borrego, S.L.; Lin, D.-W.; Kaiser, P. Isolation and Characterization of Methionine-Independent Clones from Methionine-Dependent Cancer Cells. Methods Mol. Biol. 2019, 1866, 37–48. [Google Scholar]
- Vanhamme, L.; Szpirer, C. Spontaneous and 5-azacytidine-induced revertants of methionine-dependent tumor-derived and H-ras-1-transformed cells. Exp. Cell Res. 1989, 181, 159–168. [Google Scholar] [CrossRef]
- Tisdale, M.J. Utilization of preformed and endogenously synthesized methionine by cells in tissue culture. Br. J. Cancer 1984, 49, 315–320. [Google Scholar] [CrossRef] [Green Version]
- Stipanuk, M.H. SULFUR AMINO ACID METABOLISM: Pathways for Production and Removal of Homocysteine and Cysteine. Annu. Rev. Nutr. 2004, 24, 539–577. [Google Scholar] [CrossRef] [PubMed]
- Mehrmohamadi, M.; Liu, X.; Shestov, A.A.; Locasale, J.W. Characterization of the Usage of the Serine Metabolic Network in Human Cancer. Cell Rep. 2014, 9, 1507–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verschoor, M.L.; Singh, G. Ets-1 regulates intracellular glutathione levels: Key target for resistant ovarian cancer. Mol. Cancer 2013, 12, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrego, S.L.; Kaiser, P.; University of California, Irvine. Unpublished work. 2018.
- Maddocks, O.D.K.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef] [Green Version]
- Mattaini, K.R.; Sullivan, M.R.; Vander Heiden, M.G. The importance of serine metabolism in cancer. J. Cell Biol. 2016, 214, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [Green Version]
- Hartwell, L.H.; Weinert, T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science 1989, 246, 629–634. [Google Scholar] [CrossRef] [Green Version]
- Harrison, J.C.; Haber, J.E. Surviving the Breakup: The DNA Damage Checkpoint. Annu. Rev. Genet. 2006, 40, 209–235. [Google Scholar] [CrossRef] [Green Version]
- Su, N.Y.; Flick, K.; Kaiser, P. The F-box protein Met30 is required for multiple steps in the budding yeast cell cycle. Mol. Cell. Biol. 2005, 25, 3875–3885. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, P.; Su, N.Y.; Yen, J.L.; Ouni, I.; Flick, K. The yeast ubiquitin ligase SCF-Met30: Connecting environmental and intracellular conditions to cell division. Cell Div. 2006, 1, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, E.E.; Peyraud, C.; Rouillon, A.; Surdin, K.Y.; Tyers, M.; Thomas, D. SCF Met30 -mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 2000, 19, 1613–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, P.; Flick, K.; Wittenberg, C.; Reed, S.I.I. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 2000, 102, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, P.; Sia, R.A.L.A.; Bardes, E.G.S.G.; Lew, D.J.J.; Reed, S.I.I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998, 12, 2587–2597. [Google Scholar] [CrossRef] [Green Version]
- Unger, M.W.; Hartwell, L.H. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc. Natl. Acad. Sci. USA 1976, 73, 1664–1668. [Google Scholar] [CrossRef] [Green Version]
- Ouni, I.; Flick, K.; Kaiser, P. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol. Cell. 2010, 40, 954–964. [Google Scholar] [CrossRef] [Green Version]
- Rouillon, A.; Barbey, R.; Patton, E.E.; Tyers, M.; Thomas, D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30 )complex. EMBO J. 2000, 19, 282–294. [Google Scholar] [CrossRef] [Green Version]
- Su, N.-Y.; Ouni, I.; Papagiannis, C.V.; Kaiser, P. A dominant suppressor mutation of the met30 cell cycle defect suggests regulation of the Saccharomyces cerevisiae Met4-Cbf1 transcription complex by Met32. J. Biol. Chem. 2008, 283, 11615–11624. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Dammer, E.B.; Gao, Y.; Lan, Q.; Villamil, M.A.; Duong, D.M.; Zhang, C.; Ping, L.; Lauinger, L.; Flick, K.; et al. Proteomics Links Ubiquitin Chain Topology Change to Transcription Factor Activation. Mol. Cell 2019, 76, 126–137.e7. [Google Scholar] [CrossRef]
- Lee, T.A.; Jorgensen, P.; Bognar, A.L.; Peyraud, C.; Thomas, D.; Tyers, M. Systematic Dissection of Combinatorial Control by the Met4 Transcriptional Complex. Mol. Biol. Cell 2010, 21, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Tolstykh, T.; Lee, J.; Boyd, K.; Stock, J.B.; Broach, J.R. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J. 2000, 19, 5672–5681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, R.M.; Jacobsen, S.J. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc. Natl. Acad. Sci. USA 1980, 77, 7306–7310. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, R.M. Methionine Dependence of Cancer and Aging: Methods and Protocols; Springer: New York, NY, USA, 2019; Volume 1899, ISBN 9781493987962. [Google Scholar]
- Bell, S.P.; Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002, 71, 333–374. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Rosenblatt, J.; Morgan, D.O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992, 11, 3995–4005. [Google Scholar] [CrossRef]
- Lunn, C.L.; Chrivia, J.C.; Baldassare, J.J. Activation of Cdk2/Cyclin E complexes is dependent on the origin of replication licensing factor Cdc6 in mammalian cells. Cell Cycle 2010, 9, 4533–4541. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Lapa, C.; Garcia-Velloso, M.J.; Lückerath, K.; Samnick, S.; Schreder, M.; Otero, P.R.; Schmid, J.-S.; Herrmann, K.; Knop, S.; Buck, A.K.; et al. 11C-Methionine-PET in Multiple Myeloma: A Combined Study from Two Different Institutions. Theranostics 2017, 7, 2956–2964. [Google Scholar] [CrossRef]
- Breillout, F.; Hadida, F.; Echinard-Garin, P.; Lascaux, V.; Poupon, M.F. Decreased rat rhabdomyosarcoma pulmonary metastases in response to a low methionine diet. Anticancer Res. 1987, 7, 861–867. [Google Scholar] [PubMed]
- Guo, H.; Lishko, V.K.; Herrera, H.; Groce, A.; Kubota, T.; Hoffman, R.M. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993, 53, 5676–5679. [Google Scholar] [PubMed]
- Hoshiya, Y.; Guo, H.; Kubota, T.; Inada, T.; Asanuma, F.; Yamada, Y.; Koh, J.; Kitajima, M.; Hoffman, R.M. Human tumors are methionine dependent in vivo. Anticancer Res. 1995, 15, 717–718. [Google Scholar] [PubMed]
- Komninou, D.; Leutzinger, Y.; Reddy, B.S.; Richie, J.P., Jr. Methionine Restriction Inhibits Colon Carcinogenesis. Nutr. Cancer 2006, 54, 202–208. [Google Scholar] [CrossRef]
- Tan, Y.; Xu, M.; Guo, H.; Sun, X.; Kubota, T.; Hoffman, R.M. Anticancer efficacy of methioninase in vivo. Anticancer Res. 1996, 16, 3931–3936. [Google Scholar]
- Hoffman, R.M.; Hoshiya, Y.; Guo, W. Efficacy of Methionine-Restricted Diets on Cancers In Vivo. Methots Mol. Biol. 2019, 1866, 75–81. [Google Scholar]
- Hoffman, R.M. Clinical Studies of Methionine-Restricted Diets for Cancer Patients. Methods Mol. Biol. 2019, 1866, 95–105. [Google Scholar]
- Goseki, N.; Yamazaki, S.; Shimojyu, K.; Kando, F.; Maruyama, M.; Endo, M.; Koike, M.; Takahashi, H. Synergistic Effect of Methionine-depleting Total Parenteral Nutrition with 5-Fluorouracil on Human Gastric Cancer: A Randomized, Prospective Clinical Trial. Jpn. J. Cancer Res. 1995, 86, 484–489. [Google Scholar] [CrossRef]
- Tan, Y.; Xu, M.; Tan, X.; Tan, X.; Wang, X.; Saikawa, Y.; Nagahama, T.; Sun, X.; Lenz, M.; Hoffman, R.M. Overexpression and Large-Scale Production of Recombinant L-Methionine-a-deamino-g-mercaptomethane-lyase for Novel Anticancer Therapy. Protein Expr. Purif. 1999, 9, 233–245. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Tan, Y.; Li, S.; Han, Q.; Yagi, S.; Takakura, T.; Takimoto, A.; Inagaki, K.; Kudou, D. Development of Recombinant Methioninase for Cancer Treatment. Methods Mol. Biol. 2019, 1866, 107–131. [Google Scholar]
- Kreis, W. Tumor therapy by deprivation of L-methionine: Rationale and results. Cancer Treat. Rep. 1979, 63, 1069–1072. [Google Scholar] [PubMed]
- Tisdale, M.J.; Jack, G.W.; Eridani, S. Differential sensitivity of normal and leukaemic haemopoietic cells to methionine deprivation by L-methioninase. Leuk. Res. 1983, 7, 269–277. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Murakami, T.; Kawaguchi, K.; Igarashi, K.; Tan, Y.; Li, S.; Han, Q. High Efficacy of Recombinant Methioninase on Patient-Derived Orthotopic Xenograft (PDOX) Mouse Models of Cancer. Methods Mol. Biol. 2019, 1866, 149–161. [Google Scholar] [PubMed]
- Kawaguchi, K.; Han, Q.; Li, S.; Tan, Y.; Igarashi, K.; Kiyuna, T.; Miyake, K.; Miyake, M.; Chmielowski, B.; Nelson, S.D.; et al. Targeting methionine with oral recombinant methioninase (o-rMETase) arrests a patient-derived orthotopic xenograft (PDOX) model of BRAF-V600E mutant melanoma: Implications for chronic clinical cancer therapy and prevention. Cell Cycle 2018, 17, 356–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, K.; Han, Q.; Li, S.; Tan, Y.; Igarashi, K.; Murakami, T.; Unno, M.; Hoffman, R.M. Efficacy of Recombinant Methioninase (rMETase) on Recalcitrant Cancer Patient-Derived Orthotopic Xenograft (PDOX) Mouse Models: A Review. Cells 2019, 8, 410. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Zavala, J.; Xu, M.; Zavala, J.; Hoffman, R.M. Serum methionine depletion without side effects by methioninase in metastatic breast cancer patients. Anticancer Res. 1996, 16, 3937–3942. [Google Scholar]
- Tan, Y.; Zavala, J.; Han, Q.; Xu, M.; Sun, X.; Tan, X.; Tan, X.; Magana, R.; Geller, J.; Hoffman, R.M. Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. 1998, 17, 3857–3860. [Google Scholar]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine Metabolism Regulates Maintenance and Differentiation of Human Pluripotent Stem Cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef] [Green Version]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013, 339, 222–226. [Google Scholar] [CrossRef] [Green Version]
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [PubMed]
- Miller, R.A.; Buehner, G.; Chang, Y.; Harper, J.M.; Sigler, R.; Smith-Wheelock, M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 2005, 4, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Grandison, R.C.; Piper, M.D.W.; Partridge, L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 2009, 462, 1061–1064. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.C.; Kaya, A.; Ma, S.; Kim, G.; Gerashchenko, M.V.; Yim, S.H.; Hu, Z.; Harshman, L.G.; Gladyshev, V.N. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 2014, 5, 3592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G.; et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, R.M. Methionine Restriction and Life-Span Extension. Methods Mol. Bio. 2019, 1866, 263–266. [Google Scholar]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18. [Google Scholar] [CrossRef] [Green Version]





| Cell Line | Methionine Dependence | Tumor Site |
|---|---|---|
| MDA-MB468 | Yes [7,10] | Breast |
| MCF7 | Yes [5,6,7,10] | Breast |
| MDA-MB361 | Yes [7] | Breast |
| HCC1806 | Yes [10] | Breast |
| HCC1143 | Yes [10] | Breast |
| SKBR3 | Yes [10] | Breast |
| BT-549 | Yes [10] | Breast |
| ZR-75-1 | Yes [10] | Breast |
| SUM-159 | Yes [10] | Breast |
| T47D | Yes [10] | Breast |
| W-256 | Yes [3,4] | Breast (rat) |
| MDA-MB231 | No [1], moderate [10] | Breast |
| HCC70 | No [10] | Breast |
| HCC38 | No [10] | Breast |
| SUM-149 | No [10] | Breast |
| MDA-MB231 | No [1], moderate [10] | Breast |
| BxPC3 | Yes [11] | Pancreas |
| PANC1 | No [11] | Pancreas |
| LoVo | Yes [12] | Colon |
| SK-CO-1 | Yes [6] | Colon |
| PC-3 | Yes [5,6,13] | Prostate |
| LNCaP | Moderate [13] | Prostate |
| DU145 | Moderate [5,6,13] | Prostate |
| SV80 | Yes [3] | Transformed fibroblast |
| HEK293T | Yes [11] | Transformed kidney cell |
| W18VA2 | Yes [3] | SV40 transformed human cells |
| J111 | Yes [4] | Monocytic leukemia |
| L1210 | Yes [4] | Lymphatic leukemia (mouse) |
| A2182 | Yes [5,6] | Lung |
| SK-LU-1 | Yes [5,6] | Lung |
| A549 | Moderate [6] | Lung |
| A427 | No [5,6] | Lung |
| J82 | Yes [5,6] | Bladder |
| T24 | No [5,6] | Bladder |
| 8387 | Yes [5,6] | Fibrosarcoma |
| HT1080 | Yes [5,6] | Fibrosarcoma |
| HOS | Yes [5,6] | Osteosarcoma |
| A204 | Moderate [6] | Rhabdomyosarcoma |
| A673 | Yes [5,6] | Rhabdomyosarcoma |
| SK-LMS1 | No [5,6] | Leiomyosarcoma |
| SK-N-SH | Yes [5,6] | Neuroblastoma |
| SK-N-MC | No [6] | Neuroblastoma |
| A375 | Moderate [6] | Melanoma |
| MeWo | No [6] | Melanoma |
| A172 | Moderate [5,6] | Glioblastoma |
| HeLa | Yes [6] | Cervical |
| A498 | Yes [5,6] | Kidney |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaiser, P. Methionine Dependence of Cancer. Biomolecules 2020, 10, 568. https://doi.org/10.3390/biom10040568
Kaiser P. Methionine Dependence of Cancer. Biomolecules. 2020; 10(4):568. https://doi.org/10.3390/biom10040568
Chicago/Turabian StyleKaiser, Peter. 2020. "Methionine Dependence of Cancer" Biomolecules 10, no. 4: 568. https://doi.org/10.3390/biom10040568
APA StyleKaiser, P. (2020). Methionine Dependence of Cancer. Biomolecules, 10(4), 568. https://doi.org/10.3390/biom10040568