Histamine, Metabolic Remodelling and Angiogenesis: A Systems Level Approach †
Abstract
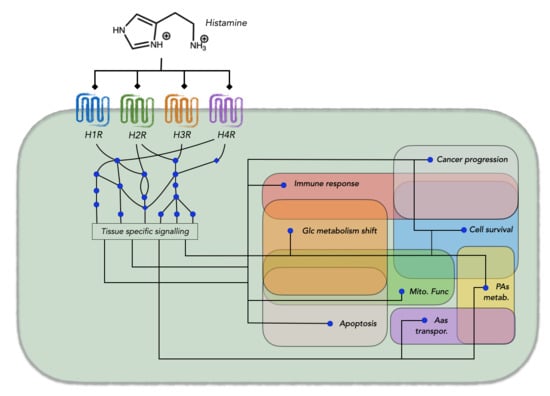
:1. Histamine Metabolism and its Connections to Other Metabolic Modules
2. Histamine and Vessel Dynamics
3. Histamine, Metabolic Reprogramming and Angiogenesis: Pathophysiological Implications
3.1. Histamine and Angiogenesis in Gestation
3.2. Histamine in Cancer
3.3. Histamine and Angiogenesis
4. A Systems Biology Approach to Histamine as a Modulator of Metabolic Reprogramming
5. Concluding Remarks and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ramos-Montañez, S.; Winkler, M.E. Biosynthesis of Histidine. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, T.; Nakamura, T.; Shibakusa, T.; Sugita, M.; Naganuma, F.; Iida, T.; Miura, Y.; Mohsen, A.; Harada, R.; Yanai, K. Insufficient Intake of l-Histidine Reduces Brain Histamine and Causes Anxiety-Like Behaviors in Male Mice. J. Nutr. 2014, 144, 1637–1641. [Google Scholar] [CrossRef]
- Panula, P.; Sundvik, M.; Karlstedt, K. Developmental roles of brain histamine. Trends Neurosci. 2014, 37, 159–168. [Google Scholar] [CrossRef]
- Stark, H. (Ed.) Histamine H 4 Receptor: A Novel Drug Target for Immunoregulation and Inflammation; Versita: Berlin, Germany, 2013; ISBN 9788376560564. [Google Scholar]
- Holeček, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [Green Version]
- Lai, T.-S.; Lin, C.-J.; Greenberg, C.S. Role of tissue transglutaminase-2 (TG2)-mediated aminylation in biological processes. Amino Acids 2017, 49, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Vowinckel, J.; Stahlberg, S.; Paulmann, N.; Bluemlein, K.; Grohmann, M.; Ralser, M.; Walther, D.J. Histaminylation of glutamine residues is a novel posttranslational modification implicated in G-protein signaling. FEBS Lett. 2012, 586, 3819–3824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, S.W.; Piper, J.; Haraldsen, G.; Oynebraten, I.; Fleckenstein, B.; Molberg, O.; Khosla, C.; Sollid, L.M. Tissue transglutaminase-mediated formation and cleavage of histamine-gliadin complexes: Biological effects and implications for celiac disease. J. Immunol. 2005, 174, 1657–1663. [Google Scholar] [CrossRef] [Green Version]
- Ohtsu, H. Histamine synthesis and lessons learned from histidine decarboxylase deficient mice. Adv. Exp. Med. Biol. 2010, 709, 21–31. [Google Scholar] [PubMed]
- Ohtsu, H. Pathophysiologic Role of Histamine: Evidence Clarified by Histidine Decarboxylase Gene Knockout Mice. Int. Arch. Allergy Immunol. 2012, 158, 2–6. [Google Scholar] [CrossRef]
- Fajardo, I.; Urdiales, J.L.; Paz, J.C.; Chavarría, T.; Sánchez-Jiménez, F.; Medina, M.Á. Histamine prevents polyamine accumulation in mouse C57.1 mast cell cultures. Eur. J. BioChem. 2001, 268, 768–773. [Google Scholar] [CrossRef] [Green Version]
- García-Faroldi, G.; Correa-Fiz, F.; Abrighach, H.; Berdasco, M.; Fraga, M.F.; Esteller, M.; Urdiales, J.L.; Sánchez-Jiménez, F.; Fajardo, I. Polyamines affect histamine synthesis during early stages of IL-3-induced bone marrow cell differentiation. J. Cell. Biochem. 2009, 108, 261–271. [Google Scholar] [CrossRef]
- García-Faroldi, G.; Rodríguez, C.E.; Urdiales, J.L.; Pérez-Pomares, J.M.; Dávila, J.C.; Pejler, G.; Sánchez-Jiménez, F.; Fajardo, I. Polyamines Are Present in Mast Cell Secretory Granules and Are Important for Granule Homeostasis. PLoS ONE 2010, 5, e15071. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Andrade, C.; Lambertos, A.; Urdiales, J.L.; Sánchez-Jiménez, F.; Peñafiel, R.; Fajardo, I. A novel role for antizyme inhibitor 2 as a regulator of serotonin and histamine biosynthesis and content in mouse mast cells. Amino Acids 2016, 48, 2411–2421. [Google Scholar] [CrossRef]
- Fernández-Reina, A.; Urdiales, J.L.; Sánchez-Jiménez, F. What We Know and What We Need to Know about Aromatic and Cationic Biogenic Amines in the Gastrointestinal Tract. Foods 2018, 7, 145. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Jiménez, F.; Ruiz-Pérez, M.V.; Urdiales, J.L.; Medina, M.A. Pharmacological potential of biogenic amine–polyamine interactions beyond neurotransmission. Br. J. Pharmacol. 2013, 170, 4–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, R.S. Chemistry and diversity of pyridoxal-5′-phosphate dependent enzymes. Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.P.; Plecko, B.; Mills, P.B.; Clayton, P.T. Disorders affecting vitamin B6 metabolism. J. Inherit. Metab. Dis. 2019, 42, 629–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panula, P.; Chazot, P.L.; Cowart, M.; Gutzmer, R.; Leurs, R.; Liu, W.L.S.; Stark, H.; Thurmond, R.L.; Haas, H.L. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors. Pharmacol. Rev. 2015, 67, 601–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, F.E.R. Advances in H1-Antihistamines. N Engl. J. Med. 2004, 351, 2203–2217. [Google Scholar] [CrossRef]
- Alexander, S.P.H.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Marrion, N.V.; Peters, J.A.; Faccenda, E.; Harding, S.D.; Pawson, A.J.; Sharman, J.L.; et al. The Concise Guide to Pharmacology 2017/18: G protein-coupled receptors. Br. J. Pharmacol. 2017, 174, S17–S129. [Google Scholar] [CrossRef] [PubMed]
- Tiligada, E.; Ennis, M. Histamine pharmacology: From Sir Henry Dale to the 21st century. Br. J. Pharmacol. 2020, 177, 469–489. [Google Scholar] [CrossRef] [Green Version]
- Moriguchi, T.; Takai, J. Histamine and histidine decarboxylase: Immunomodulatory functions and regulatory mechanisms. Genes Cells 2020, 25, 443–449. [Google Scholar] [CrossRef]
- Zhao, C.M.; Chen, D.; Yamada, H.; Dornonville De La Cour, C.; Lindström, E.; Persson, L.; Håkanson, R. Rat stomach ECL cells: Mode of activation of histidine decarboxylase. Regul. Pept. 2003, 114, 21–27. [Google Scholar] [CrossRef]
- Olmo, M.T.; Rodríguez-Agudo, D.; Medina, M.A.; Sánchez-Jiménez, F. The pest regions containing C-termini of mammalian ornithine decarboxylase and histidine decarboxylase play different roles in protein degradation. Biochem. Biophys. Res. Commun. 1999, 257, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Agudo, D.; Olmo, M.T.; Sánchez-Jiménez, F.; Medina, M.Á. Rat Histidine Decarboxylase Is a Substrate for m-Calpain in Vitro. Biochem. Biophys. Res. Commun. 2000, 271, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.V.; Fajardo, I.; Langlois, M.R.; Sánchez-Jiménez, F.; Wang, T.C. The C-terminus of rat L-histidine decarboxylase specifically inhibits enzymic activity and disrupts pyridoxal phosphate-dependent interactions with L-histidine substrate analogues. Biochem. J. 2004, 381, 769–778. [Google Scholar] [CrossRef] [Green Version]
- Furuta, K.; Nakayama, K.; Sugimoto, Y.; Ichikawa, A.; Tanaka, S. Activation of histidine decarboxylase through post-translational cleavage by caspase-9 in a mouse mastocytoma P-815. J. Biol. Chem. 2007, 282, 13438–13446. [Google Scholar] [CrossRef] [Green Version]
- Fennell, L.M.; Fleming, J. V Differential processing of mammalian l-histidine decarboxylase enzymes. Biochem. Biophys. Res. Commun. 2014, 445, 304–309. [Google Scholar] [CrossRef]
- Rodríguez-Caso, C.; Rodríguez-Agudo, D.; Moya-García, A.A.; Fajardo, I.; Medina, M.Á.; Subramaniam, V.; Sánchez-Jiménez, F. Local changes in the catalytic site of mammalian histidine decarboxylase can affect its global conformation and stability. Eur. J. Biochem. 2003, 270, 4376–4387. [Google Scholar] [CrossRef] [Green Version]
- Fleming, J.V.; Sánchez-Jiménez, F.; Moya-García, A.A.; Langlois, M.R.; Wang, T.C. Mapping of catalytically important residues in the rat l-histidine decarboxylase enzyme using bioinformatic and site-directed mutagenesis approaches. Biochem. J. 2004, 379, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moya-García, A.A.; Medina, M.Á.; Sánchez-Jiménez, F. Mammalian histidine decarboxylase: From structure to function. Bioessays 2005, 27, 57–63. [Google Scholar] [CrossRef]
- Komori, H.; Nitta, Y.; Ueno, H.; Higuchi, Y. Purification, crystallization and preliminary X-ray analysis of human histidine decarboxylase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 675–677. [Google Scholar] [CrossRef]
- Moya-García, A.A.; Ruiz-Pernía, J.; Martí, S.; Sánchez-Jiménez, F.; Tuñón, I. Analysis of the decarboxylation step in mammalian histidine decarboxylase. A computational study. J. Biol. Chem. 2008, 283, 12393–12401. [Google Scholar] [CrossRef] [Green Version]
- Caro-Astorga, J.; Fajardo, I.; Ruiz-Pérez, M.V.; Sánchez-Jiménez, F.; Urdiales, J.L. Nascent histamine induces α-synuclein and caspase-3 on human cells. Biochem. Biophys. Res. Commun. 2014, 451, 580–586. [Google Scholar] [CrossRef]
- Chen, D.; Aihara, T.; Zhao, C.M.; Håkanson, R.; Okabe, S. Differentiation of the Gastric Mucosa I. Role of histamine in control of function and integrity of oxyntic mucosa: Understanding gastric physiology through disruption of targeted genes. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Jiménez, F.; Montañez, R.; Correa-Fiz, F.; Rodríguez-Caso, C.; Urdiales, J.L.; Aldana, J.F.; Medina, M.Á. The usefulness of post-genomics tools for characterization of the amine cross-talk in mammalian cells. Biochem. Soc. Trans. 2007, 35, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Valent, P.; Akin, C.; Hartmann, K.; Nilsson, G.; Reiter, A.; Hermine, O.; Sotlar, K.; Sperr, W.R.; Escribano, L.; George, T.I.; et al. Mast cells as a unique hematopoietic lineage and cell system: From Paul Ehrlich’s visions to precision medicine concepts. Theranostics 2020, 10, 10743–10768. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Okamoto, M.; Shibue, R.; Mizuta, T.; Shibayama, T.; Yoshino, T.; Murakami, T.; Yamaguchi, M.; Tanaka, S.; Toida, T.; et al. KLF4 is required for suppression of histamine synthesis by polyamines during bone marrow-derived mast cell differentiation. PLoS ONE 2020, 15, e0229744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanerva, K.; Lappalainen, J.; Mäkitie, L.T.; Virolainen, S.; Kovanen, P.T.; Andersson, L.C. Expression of Antizyme Inhibitor 2 in Mast Cells and Role of Polyamines as Selective Regulators of Serotonin Secretion. PLoS ONE 2009, 4, e6858. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Molina, B.; Lambertos, A.; Peñafiel, R. Antizyme Inhibitors in Polyamine Metabolism and Beyond: Physiopathological Implications. Med. Sci. 2018, 6, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatsunami, K.; Ohtsu, H.; Tsuchikawa, M.; Higuchi, T.; Ishibashi, K.; Shida, A.; Shima, Y.; Nakagawa, S.; Yamauchi, K.; Yamamoto, M. Structure of the L-histidine decarboxylase gene. J. Biol. Chem. 1994, 269, 1554–1559. [Google Scholar] [CrossRef]
- Höcker, M.; Zhang, Z.; Koh, T.J.; Wang, T.C. The regulation of histidine decarboxylase gene expression. Yale J. Biol. Med. 1996, 69, 21–33. [Google Scholar] [PubMed]
- Ai, W.; Liu, Y.; Langlois, M.; Wang, T.C. Kruppel-like Factor 4 (KLF4) Represses Histidine Decarboxylase Gene Expression through an Upstream Sp1 Site and Downstream Gastrin Responsive Elements. J. Biol. Chem. 2004, 279, 8684–8693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, W.; Liu, Y.; Wang, T.C. Yin yang 1 (YY1) represses histidine decarboxylase gene expression with SREBP-1a in part through an upstream Sp1 site. Am. J. Physiol. Gastrointest Liver Physiol. 2006, 290. [Google Scholar] [CrossRef]
- Kuramasu, A.; Saito, H.; Suzuki, S.; Watanabe, T.; Ohtsu, H. Mast cell-/basophil-specific transcriptional regulation of human L-histidine decarboxylase gene by CpG methylation in the promoter region. J. Biol. Chem. 1998, 273, 31607–31614. [Google Scholar] [CrossRef] [Green Version]
- Correa-Fiz, F.; Reyes-Palomares, A.; Fajardo, I.; Melgarejo, E.; Gutiérrez, A.; García-Ranea, J.A.; Medina, M.A.; Sánchez-Jiménez, F. Regulatory cross-talk of mouse liver polyamine and methionine metabolic pathways: A systemic approach to its physiopathological consequences. Amino Acids 2012, 42, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, N. Expression of Histidine Decarboxylase and Its Roles in Inflammation. Int. J. Mol. Sci. 2019, 20, 376. [Google Scholar] [CrossRef] [Green Version]
- Schwelberger, H.G.; Ahrens, F.; Fogel, W.A.; Sánchez-Jiménez, F. Histamine metabolism. In Histamine H4 Receptor: A Novel Drug Target in Immunoregulation and Inflammation; Stark, H., Ed.; Versita: Berlin, Germany, 2013; pp. 63–102. ISBN 9788376560564. [Google Scholar]
- Horton, J.R.; Sawada, K.; Nishibori, M.; Zhang, X.; Cheng, X. Two polymorphic forms of human histamine methyltransferase: Structural, thermal, and kinetic comparisons. Structure 2001, 9, 837–849. [Google Scholar] [CrossRef] [Green Version]
- Schwelberger, H.G. Histamine N-methyltransferase (HNMT) enzyme and gene. In Histamine: Biology and Medical Aspects; Falus, A., Grosman, N., Darvas, Z., Eds.; SpringMed. Publishing: Budapest, Hungary, 2004; pp. 53–59. ISBN 9789639456396. [Google Scholar]
- Wang, L.; Yan, L.; McGuire, C.; Kozak, C.A.; Wang, M.; Kim, U.J.; Siciliano, M.; Weinshilboum, R.M. Mouse histamine N-methyltransferase: cDNA cloning, expression, gene cloning and chromosomal localization. Inflamm. Res. 2001, 50, 300–308. [Google Scholar] [CrossRef]
- Reyes-Palomares, A.; Montañez, R.; Sánchez-Jiménez, F.; Medina, M.A. A combined model of hepatic polyamine and sulfur amino acid metabolism to analyze S-adenosyl methionine availability. Amino Acids 2012, 42, 597–610. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Caso, C.; Montañez, R.; Cascante, M.; Sánchez-Jiménez, F.; Medina, M.Á. Mathematical modeling of polyamine metabolism in mammals. J. Biol. Chem. 2006, 281, 21799–21812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biebl, M.; Klocker, J.; Perkmann, R.; Kolbitsch, C.; Klingler, P.; Drasche, A.; Klingler, A.; Fraedrich, G.; Schwelberger, H.G. Heparin-induced diamine oxidase release into the circulation in pigs. Inflamm. Res. 2002, 51, S93–S94. [Google Scholar] [CrossRef] [PubMed]
- Arige, V.; Agarwal, A.; Khan, A.A.; Kalyani, A.; Natarajan, B.; Gupta, V.; Reddy, S.S.; Barthwal, M.K.; Mahapatra, N.R. Regulation of Monoamine Oxidase B Gene Expression: Key Roles for Transcription Factors Sp1, Egr1 and CREB, and microRNAs miR-300 and miR-1224. J. Mol. Biol. 2019, 431, 1127–1147. [Google Scholar] [CrossRef]
- Maslinski, C.; Fogel, W.A. Catabolism of histamine. In Histamine and Histamine Antagonists; Uvnäs, B., Ed.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 165–189. ISBN 9783642758409. [Google Scholar]
- Schwelberger, H.G.; Feurle, J.; Houen, G. New tools for studying old questions: Antibodies for human diamine oxidase. J. Neural. Transm. 2013, 120, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Fogel, W.A. Mucosal mono- and polyamine oxidase activities in digestive tract are distributed complementary to diamine oxidase. J. Neural. Transm. Suppl. 1990, 32, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yan, L.-J. Protein Oxidative Modifications: Beneficial Roles in Disease and Health. J. Biochem. Pharmacol. Res 2013, 1, 15–26. [Google Scholar] [PubMed]
- Nagy, J.A.; Benjamin, L.; Zeng, H.; Dvorak, A.M.; Dvorak, H.F. Vascular permeability, vascular hyperpermeability and angiogenesis. Angiogenesis 2008, 11, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Halpern, B.N.; Guillaumat, L.; Cruchaud, S. Action of histamine and synthetic antihistamines on the capillary permeability of the blood-brain barrier. Sem Hop 1948, 24, 2564. [Google Scholar]
- Miles, A.A. Increased permeability of skin capillaries produced by histamine in guinea-pigs. J. Physiol. 1951, 114, 35. [Google Scholar]
- Gözsy, B.; Kátó, L. Changes in permeability of the skin capillaries of rats after histamine depletion with 48/80, dextran or egg white. J. Physiol. 1957, 139, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Majno, G.; Palade, G.E.; Schoefl, G.I. Studies on Inflammation II. The Site of Action of Histamine and Serotonin along the Vascular Tree: A Topographic Study. J. Biophys. Biochem. Cytol. 1961, 11, 607–626. [Google Scholar] [CrossRef] [Green Version]
- Fredholm, B.B.; Meng, H.C.; Oberg, B.; Rosell, S. Vascular and metabolic effects of histamine and compound 48/80 in subcutaneous adipose tissue. Br. J. Pharmacol. 1968, 34, 197–198. [Google Scholar]
- Buckley, I.K.; Ryan, G.B. Increased vascular permeability. The effect of histamine and serotonin on rat mesenteric blood vessels in vivo. Am. J. Pathol. 1969, 55, 329–347. [Google Scholar] [PubMed]
- van Nieuw Amerongen, G.P.; Draijer, R.; Vermeer, M.A.; van Hinsbergh, V.W.M. Transient and Prolonged Increase in Endothelial Permeability Induced by Histamine and Thrombin. Circ. Res. 1998, 83, 1115–1123. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Utoguchi, N.; Makimoto, H.; Mizuguchi, H.; Nakagawa, S.; Mayumi, T. Different Reactions of Aortic and Venular Endothelial Cell Monolayers to Histamine on Macromolecular Permeability: Role of cAMP, Cytosolic Ca2+ and F-actin. Inflammation 1999, 23, 87–97. [Google Scholar] [CrossRef]
- Guo, M.; Breslin, J.W.; Wu, M.H.; Gottardi, C.J.; Yuan, S.Y. VE-cadherin and β-catenin binding dynamics during histamine-induced endothelial hyperpermeability. Am J. Physiol. 2008, 294, C977–C984. [Google Scholar] [CrossRef] [Green Version]
- Rozenberg, I.; Sluka, S.H.M.; Rohrer, L.; Hofmann, J.; Becher, B.; Akhmedov, A.; Soliz, J.; Mocharla, P.; Borén, J.; Johansen, P.; et al. Histamine H1 Receptor Promotes Atherosclerotic Lesion Formation by Increasing Vascular Permeability for Low-Density Lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlstedt, K.; Jin, C.; Panula, P. Expression of histamine receptor genes HRH3 and HRH4 in rat brain endothelial cells. Br. J. Pharmacol. 2013, 170, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Adderley, S.P.; Zhang, X.E.; Breslin, J.W. Involvement of the H1 Histamine Receptor, p38 MAP Kinase, Myosin Light Chains Kinase, and Rho/ROCK in Histamine-Induced Endothelial Barrier Dysfunction. Microcirculation 2015, 22, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Ashina, K.; Tsubosaka, Y.; Nakamura, T.; Omori, K.; Kobayashi, K.; Hori, M.; Ozaki, H.; Murata, T. Histamine Induces Vascular Hyperpermeability by Increasing Blood Flow and Endothelial Barrier Disruption In Vivo. PLoS ONE 2015, 10, e0132367. [Google Scholar] [CrossRef]
- Mikelis, C.M.; Simaan, M.; Ando, K.; Fukuhara, S.; Sakurai, A.; Amornphimoltham, P.; Masedunskas, A.; Weigert, R.; Chavakis, T.; Adams, R.H.; et al. RhoA and ROCK mediate histamine-induced vascular leakage and anaphylactic shock. Nat. Commun. 2015, 6, 6725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kugelmann, D.; Rotkopf, L.T.; Radeva, M.Y.; Garcia-Ponce, A.; Walter, E.; Waschke, J. Histamine causes endothelial barrier disruption via Ca2+-mediated RhoA activation and tension at adherens junctions. Sci. Rep. 2018, 8, 13229. [Google Scholar] [CrossRef]
- Si, H.; Wang, J.; Meininger, C.J.; Peng, X.; Zawieja, D.C.; Zhang, S.L. Ca2+ release-activated Ca2+ channels are responsible for histamine-induced Ca2+ entry, permeability increase, and interleukin synthesis in lymphatic endothelial cells. Am. J. Physiol. Circ. Physiol. 2020, 318, H1283–H1295. [Google Scholar] [CrossRef]
- Grimsey, N.J.; Lin, Y.; Narala, R.; Rada, C.C.; Mejia-Pena, H.; Trejo, J. G protein–coupled receptors activate p38 MAPK via a non-canonical TAB1–TAB2– and TAB1–TAB3–dependent pathway in endothelial cells. J. Biol. Chem. 2019, 294, 5867–5878. [Google Scholar] [CrossRef] [Green Version]
- Melgarejo, E.; Medina, M.Á.; Sánchez-Jiménez, F.; Botana, L.M.; Domínguez, M.; Escribano, L.; Orfao, A.; Urdiales, J.L. (-)-Epigallocatechin-3-gallate interferes with mast cell adhesiveness, migration and its potential to recruit monocytes. Cell Mol. Life Sci. 2007, 64, 2690–2701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melgarejo, E.; Medina, M.Á.; Sánchez-Jiménez, F.; Urdiales, J.L. Epigallocatechin gallate reduces human monocyte mobility and adhesion in vitro. Br. J. Pharmacol. 2009, 158, 1705–1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Garcia, C.; Lopez-Contreras, A.J.; Cremades, A.; Castells, M.T.; Peñafiel, R. Transcriptomic Analysis of Polyamine-Related Genes and Polyamine Levels in Placenta, Yolk Sac and Fetus During the Second Half of Mouse Pregnancy. Placenta 2009, 30, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ramirez, V.D. Dibutyryl cyclic adenosine monophosphate stimulates in vitro luteinizing hormone-releasing hormone release only from median eminence derived from ovariectomized, estradiol-priMed. rats. Brain Res. 1985, 342, 154–157. [Google Scholar] [CrossRef]
- Soares, M.J.; Iqbal, K.; Kozai, K. Hypoxia and Placental Development. Birth Defects Res. 1309, 109, 1309–1329. [Google Scholar] [CrossRef]
- Maintz, L.; Schwarzer, V.; Bieber, T.; Ven, K.; van der Novak, N. Effects of histamine and diamine oxidase activities on pregnancy: A critical review. Hum. Reprod. Update 2008, 14, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Crouse, M.S.; Greseth, N.P.; McLean, K.J.; Crosswhite, M.R.; Pereira, N.N.; Ward, A.K.; Reynolds, L.P.; Dahlen, C.R.; Neville, B.W.; Borowicz, P.P.; et al. Maternal nutrition and stage of early pregnancy in beef heifers: Impacts on hexose and AA concentrations in maternal and fetal fluids. J. Anim Sci. 2019, 97, 1296–1316. [Google Scholar] [CrossRef] [PubMed]
- Pyzlak, M.; Szewczyk, G.; Szukiewicz, D.; Szczesniak, A. Histamine influence on apoptosis in trophoblast cell cultures. Inflamm. Res. 2010, 59, 213–215. [Google Scholar] [CrossRef]
- Komi, D.E.A.; Shafaghat, F.; Haidl, G. Significance of mast cells in spermatogenesis, implantation, pregnancy, and abortion: Cross talk and molecular mechanisms. Am. J. Reprod Immunol. 2020, 83, e13228. [Google Scholar] [CrossRef] [Green Version]
- Faas, M.M.; Vos, P. De Innate immune cells in the placental bed in healthy pregnancy and preeclampsia. Placenta 2018, 69, 125–133. [Google Scholar] [CrossRef]
- Medina, M.Á.; Quesada, A.R.; Núñez de Castro, I.; Sánchez-Jiménez, F. Histamine, polyamines, and cancer. Biochem. Pharmacol. 1999, 57, 1341–1344. [Google Scholar] [CrossRef]
- Medina, V.A.; Coruzzi, G.; Lamas, D.J.M.; Massari, N.; Adami, M.; Levi-Schaffer, F.; Ben-Zimra, M.; Schwelberger, H.; Rivera, E.S. Histamine in cancer. In Histamine H4 Receptor: A Novel Drug Target in Immunoregulation and Inflammation; Versita: Berlin, Germany; Volume 9788376560, pp. 259–308. ISBN 9788376560.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krauth, M.-T.T.; Agis, H.; Aichberger, K.J.; Simonitsch-Klupp, I.; Müllauer, L.; Mayerhofer, M.; Böhm, A.; Horny, H.-P.P.; Valent, P. Immunohistochemical detection of histidine decarboxylase in neoplastic mast cells in patients with systemic mastocytosis. Hum. Pathol. 2006, 37, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Rivera, E.S.; Cricco, G.P.; Engel, N.I.; Fitzsimons, C.P.; Martín, G.A.; Bergoc, R.M. Histamine as an autocrine growth factor: An unusual role for a widespread mediator. Semin. Cancer Biol. 2000, 10, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Höcker, M.; Rosenberg, I.; Xavier, R.; Henihan, R.J.; Wiedenmann, B.; Rosewicz, S.; Podolsky, D.K.; Wang, T.C. Oxidative Stress Activates the Human Histidine Decarboxylase Promoter in AGS Gastric Cancer Cells*. J. Biol. Chem. 1998, 273, 23046–23054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urdiales, J.L.; Mates, J.M.; Núñez de Castro, I.; Sánchez-Jiménez, F. Chlorpheniramine inhibits the ornithine decarboxylase induction of Ehrlich carcinoma growing in vivo. FEBS Lett. 1992, 305, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Medina, V.A.; Rivera, E.S. Histamine receptors and cancer pharmacology. Br. J. Pharmacol. 2010, 161, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Martinel Lamas, D.J.; Croci, M.; Carabajal, E.; Crescenti, E.J.V.; Sambuco, L.; Massari, N.A.; Bergoc, R.M.; Rivera, E.S.; Medina, V.A. Therapeutic potential of histamine H4 receptor agonists in triple-negative human breast cancer experimental model. Br. J. Pharmacol. 2013, 170, 188–199. [Google Scholar] [CrossRef] [Green Version]
- Massari, N.A.; Medina, V.A.; Martinel Lamas, D.J.; Cricco, G.P.; Croci, M.; Sambuco, L.; Bergoc, R.M.; Rivera, E.S. Role of H4 receptor in histamine-mediated responses in human melanoma. Melanoma Res. 2011, 21, 395–404. [Google Scholar] [CrossRef]
- Pós, Z.; Sáfrány, G.; Müller, K.; Tóth, S.; Falus, A.; Hegyesi, H. Phenotypic Profiling of Engineered Mouse Melanomas with Manipulated Histamine Production Identifies Histamine H2 Receptor and Rho-C as Histamine-Regulated Melanoma Progression Markers. Cancer Res 2005, 65, 4458–4466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suarez-Carmona, M.; Lesage, J.; Cataldo, D.; Gilles, C. EMT and inflammation: Inseparable actors of cancer progression. Mol. Oncol. 2017, 11, 805–823. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P. Angiogenesis in life, disease and medicine. Nature 2005, 438, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Caso, L.; Reyes-Palomares, A.; Sánchez-Jiménez, F.; Quesada, A.R.; Medina, M.Á. What is known on angiogenesis-related rare diseases? A systematic review of literature. J. Cell Mol. Med. 2012, 16, 2872–2893. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.A.; Simpson, J.G. Ciba Foundation Symposium 100—Development of the Vascular System. Novartis Found. Symp. 2008, 100, 120–131. [Google Scholar] [CrossRef]
- Barnhill, R.L.; Ryan, T.J. Biochemical Modulation of Angiogenesis in the Chorioallantoic Membrane of the Chick Embryo. J. Investig. Dermatol. 1983, 81, 485–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marks, R.M.; Roche, W.R.; Czerniecki, M.; Penny, R.; Nelson, D.S. Mast cell granules cause proliferation of human microvascular endothelial cells. Lab. Investig. 1986, 55, 289–294. [Google Scholar]
- Thompson, W.D.; Brown, F.I. Quantitation of histamine-induced angiogenesis in the chick chorioallantoic membrane: Mode of action of histamine is indirect. Int. J. Microcirc. Clin. Exp. 1987, 6, 343–357. [Google Scholar]
- Sörbo, J.; Jakobsson, A.; Norrby, K. Mast-cell histamine is angiogenic through receptors for histamine1 and histamine2. Int. J. Exp. Pathol. 1994, 75, 43–50. [Google Scholar] [PubMed]
- Qin, L.; Zhao, D.; Xu, J.; Ren, X.; Terwilliger, E.F.; Parangi, S.; Lawler, J.; Dvorak, H.F.; Zeng, H. The vascular permeabilizing factors histamine and serotonin induce angiogenesis through TR3/Nur77 and subsequently truncate it through thrombospondin-1. Blood 2013, 121, 2154–2164. [Google Scholar] [CrossRef] [Green Version]
- Norrby, K. Evidence of a dual role of endogenous histamine in angiogenesis. Int. J. Exp. Pathol. 1995, 76, 87–92. [Google Scholar]
- Norrby, K. Mast cells and angiogenesis. APMIS 2002, 110, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Varricchi, G.; Rossi, F.W.; Galdiero, M.R.; Granata, F.; Criscuolo, G.; Spadaro, G.; de Paulis, A.; Marone, G. Physiological Roles of Mast Cells: Collegium Internationale Allergologicum Update 2019. Int. Arch. Allergy Immunol 2019, 179, 247–261. [Google Scholar] [CrossRef]
- Komi, D.E.A.; Wöhrl, S.; Bielory, L. Mast Cell Biology at Molecular Level: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2020, 58, 342–365. [Google Scholar] [CrossRef] [PubMed]
- Laakkonen, J.P.; Lappalainen, J.P.; Theelen, T.L.; Toivanen, P.I.; Nieminen, T.; Jauhiainen, S.; Kaikkonen, M.U.; Sluimer, J.C.; Ylä-Herttuala, S. Differential regulation of angiogenic cellular processes and claudin-5 by histamine and VEGF via PI3K-signaling, transcription factor SNAI2 and interleukin-8. Angiogenesis 2017, 20, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Wang, C.; Pan, R.; Gao, X.; Wei, Z.; Xia, Y.; Dai, Y. Histamine synergistically promotes bFGF-induced angiogenesis by enhancing VEGF production via H1 receptor. J. Cell. Biochem. 2013, 114, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Natori, T.; Sata, M.; Nagai, R.; Makuuchi, M. Cimetidine inhibits angiogenesis and suppresses tumor growth. BioMed. Pharmacother. 2005, 59, 56–60. [Google Scholar] [CrossRef]
- Medina, M.Á. Systems biology for molecular life sciences and its impact in biomedicine. Cell Mol. Life Sci. 2013, 70, 1035–1053. [Google Scholar] [CrossRef]
- Ocaña, M.C.; Martínez-Poveda, B.; Quesada, A.R.; Medina, M.Á. Metabolism within the tumor microenvironment and its implication on cancer progression: An ongoing therapeutic target. Med. Res. Rev. 2019, 39, 70–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuperstein, I.; Bonnet, E.; Nguyen, H.-A.; Cohen, D.; Viara, E.; Grieco, L.; Fourquet, S.; Calzone, L.; Russo, C.; Kondratova, M.; et al. Atlas of Cancer Signalling Network: A systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis 2015, 4, e160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodchenkov, I.; Babur, O.; Luna, A.; Aksoy, B.A.; Wong, J.V.; Fong, D.; Franz, M.; Siper, M.C.; Cheung, M.; Wrana, M.; et al. Pathway Commons 2019 Update: Integration, analysis and exploration of pathway data. Nucleic. Acids Res. 2019, 48, D489–D497. [Google Scholar] [CrossRef] [Green Version]
- Hornbeck, P.V.; Chabra, I.; Kornhauser, J.M.; Skrzypek, E.; Zhang, B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004, 4, 1551–1561. [Google Scholar] [CrossRef]
- Perfetto, L.; Briganti, L.; Calderone, A.; Perpetuini, A.C.; Iannuccelli, M.; Langone, F.; Licata, L.; Marinkovic, M.; Mattioni, A.; Pavlidou, T.; et al. SIGNOR: A database of causal relationships between biological entities. Nucleic Acids Res. 2016, 44, D548–D554. [Google Scholar] [CrossRef]
- Cui, Q.; Ma, Y.; Jaramillo, M.; Bari, H.; Awan, A.; Yang, S.; Zhang, S.; Liu, L.; Lu, M.; O’Connor-Mccourt, M.; et al. A map of human cancer signaling. Mol. Syst. Biol. 2007, 3. [Google Scholar] [CrossRef]
- Türei, D.; Korcsmáros, T.; Saez-Rodriguez, J. OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef]
- Ruiz-Perez, M.; Sanchez-Jimenez, F.; Alonso, F.; Segura, J.; Marquez, J.; Medina, M. Glutamine, Glucose and other Fuels for Cancer. Curr. Pharm. Des. 2014, 20, 2557–2579. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ryoo, M.; Skay, A.; Tomasi, I.; Giordano, P.; Mato, J.M.; Lu, S.C. Polyamine and methionine adenosyltransferase 2A crosstalk in human colon and liver cancer. Exp. Cell Res. 2013, 319, 1902–1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, M.Á.; Urdiales, J.L.; Rodríguez-Caso, C.; Ramírez, F.J.; Sánchez-Jiménez, F. Biogenic Amines and Polyamines: Similar Biochemistry for Different Physiological Missions and Biomedical Applications. Crit. Rev. Biochem. Mol. Biol. 2008, 38, 23–59. [Google Scholar] [CrossRef] [PubMed]
- Abrighach, H.; Fajardo, I.; Sánchez-Jiménez, F.; Urdiales, J.L. Exploring polyamine regulation by nascent histamine in a human-transfected cell model. Amino Acids 2010, 38, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Jimenez, F.; Reyes-Palomares, A.; Moya-Garcia, A.; Ranea, J.; Medina, M. Biocomputational Resources Useful for Drug Discovery Against Compartmentalized Targets. Curr. Pharm. Des. 2014, 20, 293–300. [Google Scholar] [CrossRef]
- Pezzuto, A.; Carico, E. Role of HIF-1 in Cancer Progression: Novel Insights. A Review. Curr. Mol. Med. 2019, 18, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Pino-Ángeles, A.; Reyes-Palomares, A.; Melgarejo, E.; Sánchez-Jiménez, F. Histamine: An undercover agent in multiple rare diseases? J. Cell Mol. Med. 2012, 16, 1947–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| H1 Receptor | H2 Receptor | H3 Receptor | H4 Receptor | |
|---|---|---|---|---|
| HGNC | HRH1 | HRH2 | HRH3 | HRH4 |
| UniprotKB | P35367 | P25021 | Q9Y5N1 | Q9H3N8 |
| Mass (kDa) | 55.7 | 40.1–44.5 (2 isoforms) | 36.4–49.6 (7 isoforms) | 34.5–44.5 (2 isoforms) |
| Binding affinity | Low (2.5 × 10−5 M) | Low (7.9 × 10−6 M) | High (6.3 × 10−9 M) | High (7.9 × 10−9 M) |
| Cell/tissue expression | Ubiquitous, brain, smooth muscle, epithelial and endothelial cells, immune cells, hepatocytes and chondrocytes | Ubiquitous, gastric-mucosa parietal cells, smooth muscle, heart, epithelial and endothelial cells, immune cells, hepatocytes and chondrocytes | High expression on histaminergic neurons | High expression on bone marrow and peripheral hematopoietic cells |
| Gα protein coupling | Gαq/11 | Gαs | Gαi/o | Gαi/o |
| Signalling pathways | PLC activation, increase of Ca2+, PKC activation, NOS activation, increase of cGMP, cAMP accumulation (via Gβγ subunits) | PKA activation, increase of cAMP, PLC activation, increase of Ca2+ | Decrease of cAMP, inhibition of Ca2+ channels, stimulation of MAP kinase phosphorylation | Decrease of cAMP, inhibition of Ca2+ channels, stimulation of MAP kinase phosphorylation |
| Primary functions | Immediate allergic response. Inflammatory response | Gastric acid secretion. Suppression of immune cells. Inflammatory response | Regulation of arousal and cognition. Control of inflammatory response | Allergic and inflammatory responses. Immune cell chemotaxis |
| Protein | HGNC | UniprotKB | Biological Function | Metabolism Remodelling |
|---|---|---|---|---|
| hexokinase 1 | HK1 | P19367 | Key glycolytic enzyme responsible of hexose phosphorylation, also involved in release of mitochondrial pro-apoptosis elements. | |
| hexokinase 2 | HK2 | P52789 | Key glycolytic enzyme responsible of hexose phosphorylation | Glycolysis |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | PFKFB3 | Q16875 | Key enzyme for glycolysis regulation. Proposed as a marker to distinguish between induced-pluripotent stem cells and cancer stem cells. Its expression is modified by hypoxia | |
| pyruvate dehydrogenase kinase 1 | PDK1 | O15530 | It activates by phosphorylation targets such as AKT1, PRKACA, involved in glucose and nitrogen uptake y storage. It can inhibit TGF-β signalling, as well as activate NF-kB in macrophages and calcium movements in mast cells. Regulator of key nutrient receptor in thymocytes, and essential for mobility of vascular endothelial cells. | TCA cycle |
| cytochrome c | CYCS | P99999 | Electron carrier protein that plays a role in the mitochondrial-associated mechanism of apoptosis | OXPHOS |
| phosphoglycerate mutase 1 | PGAM1 | P18669 | Glycolytic enzyme described as a promising target for diagnosis and therapy of cancer | Pentose phosphate pathway |
| SLC7A8 amino acid transporter light chain, L system | LAT2 | Q9UHI5 | Neutral amino acid cytosolic exchanger. It is involved in glutamine-dependent mTOR activation to promote glycolysis in cancer cells. | Amino acid metabolism |
| solute carrier family 38 member 2 | SLC38A2 | Q96QD8 | It has glutamine as a ligand, and is involved in cellular response to starvation, regulation of gene expression and splicing, and cellular response to stress. | |
| solute carrier family 7 member 1 | SLC7A1 | P30825 | It accepts L-Arg, L-ornithine, L- His and L- Lys as substrates. | |
| potassium inwardly rectifying channel subfamily J member 11 | KCNJ11 | Q14654 | It acts as a transmembrane transport system and an ankyrin-binding protein. It is Involved in cardiac muscle function, ischemia response and glucose homeostasis. | Polyamine metabolism |
| spermidine/spermine N1-acetyltransferase 1 | SAT1 | P21673 | Key enzyme for polyamine degradation. Highly regulated. | |
| spermine synthase | SMS | P52788 | Enzyme responsible of spermine synthesis from spermidine and decarboxylated S-adenosylmethionine. Diminished activity is related to Snyder-Robinson syndrome. | |
| Fas cell surface death receptor | FAS | P25445 | Key element for extrinsic apoptosis pathway. Related to regulation of immune response. | Lipid synthesis |
| amyloid beta precursor protein binding family B member 1 | APBB1 | PO00213 | Transcription coregulator related to histone postranslational modifications, and regulation of many key elements for cell division and apoptosis. | Nucleic acid metabolism |
| dihydrofolate reductase | DHFR | P00374 | Key element for biomolecular methylations important for DNA synthesis and gene expression, among many other processes. | |
| ankyrin 1 | ANK1 | P16157 | Structural protein related to cytoskeletal remodelling, and organelle organization. | Metabolic-signaling pathways |
| eukaryotic translation initiation factor 2 alpha kinase 2 | EIF2AK2 | P19525 | Protein kinase acting as an inhibitor of viral infection via the integrated stress response. Also involved in regulation of apoptosis and cell proliferation, and inflammatory response. | |
| hypoxia inducible factor 1 subunit alpha | HIF1A | Q16665 | Under hypoxia, it activates a plethora of genes, involved in embryonic vascularization and tumour angiogenesis. Also related to response to virus infections, including SARS-CoV-2. | |
| mechanistic target of rapamycin kinase | MTOR | P42345 | Central regulator of cellular metabolism, growth and survival in response to hormones, growth factors, nutrients, energy and stress signals. | |
| Endonuclease 8-like 1 | NEIL1 | Q96F14 | Involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. | |
| Plasminogen | PLG | P00747 | Plasmin precursor. Plasmin acts as a proteolytic factor in a variety of other processes including ovulation, embryonic development, tissue remodelling, tumour invasion, and inflammation. | |
| protein kinase cAMP-activated catalytic subunit alpha | PRKACA | P17612 | This kinase is involved in many processes related to fuel (glucose and lipid) metabolism, cell differentiation of different cell-types, and immune cells responses, including inflammation. When activated inhibits the antiproliferative and antiinvasive effect of difluoromethylornithine (an inhibitor of polyamine synthesis). | |
| protein kinase cAMP-dependent type I regulatory subunit alpha | PRKAR1A | P10644 | Subunit responsible of the regulation of cAMP-dependent protein kinase, whose properties are briefly described above. | |
| von Hippel-Lindau tumor suppressor | VHL | P40337 | Involved in the ubiquitination and subsequent proteasomal degradation of proteins. It is involved in transcriptional repression through interactions with H1F1A, HIF1AN and histone deacetylases. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moya-García, A.A.; Pino-Ángeles, A.; Sánchez-Jiménez, F.; Urdiales, J.L.; Medina, M.Á. Histamine, Metabolic Remodelling and Angiogenesis: A Systems Level Approach. Biomolecules 2021, 11, 415. https://doi.org/10.3390/biom11030415
Moya-García AA, Pino-Ángeles A, Sánchez-Jiménez F, Urdiales JL, Medina MÁ. Histamine, Metabolic Remodelling and Angiogenesis: A Systems Level Approach. Biomolecules. 2021; 11(3):415. https://doi.org/10.3390/biom11030415
Chicago/Turabian StyleMoya-García, Aurelio A., Almudena Pino-Ángeles, Francisca Sánchez-Jiménez, José Luis Urdiales, and Miguel Ángel Medina. 2021. "Histamine, Metabolic Remodelling and Angiogenesis: A Systems Level Approach" Biomolecules 11, no. 3: 415. https://doi.org/10.3390/biom11030415
APA StyleMoya-García, A. A., Pino-Ángeles, A., Sánchez-Jiménez, F., Urdiales, J. L., & Medina, M. Á. (2021). Histamine, Metabolic Remodelling and Angiogenesis: A Systems Level Approach. Biomolecules, 11(3), 415. https://doi.org/10.3390/biom11030415