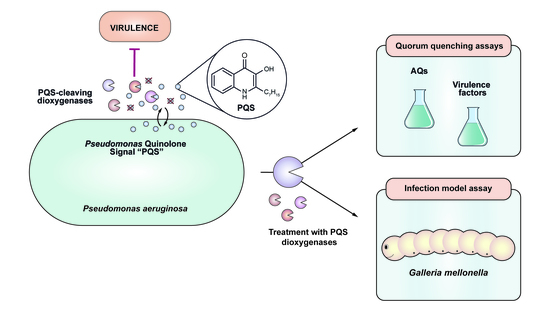
Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS): A Comparison between Naturally Occurring and Engineered PQS-Cleaving Dioxygenases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Strains, HQD Enzymes, and Plasmids
2.3. Heterologous Production and Purification of Dioxygenases
2.4. Enzyme Assay
2.5. Cultivation of P. aeruginosa in the Presence of Quorum Quenching Enzymes
2.6. Determination of AQ Levels
2.7. Quantification of Virulence Factors
2.8. Galleria mellonella Infection Model
2.9. Statistical Analysis
3. Results
3.1. Catalytic Half-Lives of PQS Dioxygenases
3.2. PQS Dioxygenases in Contrast to HodC Are Not Inactivated by P. aeruginosa Exoproducts
3.3. PQS Dioxygenases from N. farcinica and S. bingchenggensis Quench AQ and Virulence Factor Production
3.4. PQS Dioxygenase from S. bingchenggensis, as Well as Small Molecule Inhibitors of AQ Biosynthesis, Enhance G. mellonella Survival upon Infection with P. aeruginosa PAO1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal—Response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Nadal Jimenez, P.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007, 14, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Bru, J.L.; Rawson, B.; Trinh, C.; Whiteson, K.; Høyland-Kroghsbo, N.M.; Siryaporn, A. PQS produced by the Pseudomonas aeruginosa stress response repels swarms away from bacteriophage and antibiotics. J. Bacteriol. 2019, 201, e00383-19. [Google Scholar] [CrossRef] [Green Version]
- Reen, F.J.; Mooij, M.J.; Holcombe, L.J.; McSweeney, C.M.; McGlacken, G.P.; Morrissey, J.P.; O’Gara, F. The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 2011, 77, 413–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazan, R.; Que, Y.A.; Maura, D.; Strobel, B.; Majcherczyk, P.A.; Hopper, L.R.; Wilbur, D.J.; Hreha, T.N.; Barquera, B.; Rahme, L.G. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 2016, 26, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, G.F.; Park, J.; Janda, K.D. Bacterial quorum sensing: A new target for anti-infective immunotherapy. Expert Opin. Biol. Ther. 2008, 8, 719–724. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2015, 40, 86–116. [Google Scholar] [CrossRef]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2014, 201, 2–14. [Google Scholar] [CrossRef]
- Wullich, S.C.; Arranz San Martín, A.; Fetzner, S. Definition of an α/β-hydrolase fold subfamily comprising Pseudomonas quinolone signal cleaving dioxygenases. Appl. Environ. Microbiol. 2020, 86, e00279-20. [Google Scholar] [CrossRef] [PubMed]
- Pustelny, C.; Albers, A.; Büldt-Karentzopoulos, K.; Parschat, K.; Chhabra, S.R.; Cámara, M.; Williams, P.; Fetzner, S. Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 2009, 16, 1259–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birmes, F.S.; Säring, R.; Hauke, M.C.; Ritzmann, N.H.; Drees, S.L.; Daniel, J.; Treffon, J.; Liebau, E.; Kahl, B.C.; Fetzner, S. Interference with Pseudomonas aeruginosa quorum sensing and virulence by the mycobacterial PQS dioxygenase AqdC in combination with the N-acylhomoserine lactone lactonase QsdA. Infect. Immun. 2019, 87, e00278-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, C.; Birmes, F.S.; Rückert, C.; Kalinowski, J.; Fetzner, S. Rhodococcus erythropolis BG43 genes mediating Pseudomonas aeruginosa quinolone signal degradation and virulence factor attenuation. Appl. Environ. Microbiol. 2015, 81, 7720–7729. [Google Scholar] [CrossRef] [Green Version]
- Birmes, F.S.; Wolf, T.; Kohl, T.A.; Rüger, K.; Bange, F.; Kalinowski, J.; Fetzner, S. Mycobacterium abscessus subsp. abscessus is capable of degrading Pseudomonas aeruginosa quinolone signals. Front. Microbiol. 2017, 8, 339. [Google Scholar] [CrossRef] [PubMed]
- Wullich, S.C.; Wijma, H.J.; Janssen, D.B.; Fetzner, S. Stabilizing AqdC, a Pseudomonas quinolone signal-cleaving dioxygenase from Mycobacteria, by FRESCO-based protein engineering. ChemBioChem 2020, 22, 733–742. [Google Scholar] [CrossRef]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; De Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef]
- Storz, M.P.; Allegretta, G.; Kirsch, B.; Empting, M.; Hartmann, R.W. From in vitro to in cellulo: Structure-activity relationship of (2-nitrophenyl)methanol derivatives as inhibitors of PqsD in Pseudomonas aeruginosa. Org. Biomol. Chem. 2014, 12, 6094–6104. [Google Scholar] [CrossRef] [Green Version]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef] [Green Version]
- Tettmann, B.; Niewerth, C.; Kirschhöfer, F.; Neidig, A.; Dötsch, A.; Brenner-Weiss, G.; Fetzner, S.; Overhage, J. Enzyme-mediated quenching of the Pseudomonas quinolone signal (PQS) promotes biofilm formation of Pseudomonas aeruginosa by increasing iron availability. Front. Microbiol. 2016, 7, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two-anthranilate synthase and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [Green Version]
- Stintzi, A.; Cornelis, P.; Hohnadel, D.; Meyer, J.M.; Dean, C.; Poole, K.; Kourambas, S.; Krishnapillai, V. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 1996, 142, 1181–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, S.; Gdynia, A.; Tielen, P.; Rosenau, F.; Jaeger, K.E. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J. Bacteriol. 2007, 189, 6695–6703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toder, D.S.; Gambello, M.J.; Iglewski, B.H. Pseudomonas aeruginosa LasA: A second elastase under the transcriptional control of lasR. Mol. Microbiol. 1991, 5, 2003–2010. [Google Scholar] [CrossRef]
- Patidar, S.K.; Kim, S.H.; Kim, J.H.; Park, J.; Park, B.S.; Han, M.S. Pelagibaca bermudensis promotes biofuel competence of Tetraselmis striata in a broad range of abiotic stressors: Dynamics of quorum-sensing precursors and strategic improvement in lipid productivity. Biotechnol. Biofuels 2018, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Coulon, P.M.L.; Zlosnik, J.E.A.; Déziel, E. Presence of the Hmq system and production of 4-hydroxy-3-methyl-2-alkylquinolines are heterogeneously distributed between Burkholderia cepacia complex species and more prevalent among environmental than clinical isolates. Microbiol. Spectr. 2021, 9, e00127-21. [Google Scholar] [CrossRef]
- Collier, D.N.; Anderson, L.; McKnight, S.L.; Noah, T.L.; Knowles, M.; Boucher, R.; Schwab, U.; Gilligan, P.; Pesci, E.C. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 2002, 215, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.Y.; Hoke, T.; Seravalli, J.; Switzer, B.L.; Bavitz, M.; Fliege, J.D.; Murphy, P.J.; Britigan, B.E. Pseudomonas aeruginosa signal induces oxidative stress and inhibits heme oxygenase-1 expression in lung epithelial cells. Infect. Immun. 2017, 85, e00176-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, H.L.; Halliday, N.; Barrett, D.A.; Williams, P.; Forrester, D.L.; Peckham, D.; Williams, K.; Smyth, A.R.; Honeybourne, D.L.; Whitehouse, J.; et al. Diagnostic and prognostic significance of systemic alkyl quinolones for P. aeruginosa in cystic fibrosis: A longitudinal study. J. Cyst. Fibros. 2017, 16, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Que, Y.-A.; Hazan, R.; Ryan, C.M.; Milot, S.; Lépine, F.; Lydon, M.; Rahme, L.G. Production of Pseudomonas aeruginosa intercellular small signaling molecules in human burn wounds. J. Pathog. 2011, 2011, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Reyes, S.; Soberón-Chávez, G.; Cocotl-Yanez, M. The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein. J. Med. Microbiol. 2020, 69, 25–34. [Google Scholar] [CrossRef]
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [Green Version]






| Protein | Source Organism | ENA ID | Expression Plasmid | Reference |
|---|---|---|---|---|
| HodC | Arthrobacter sp. Rue61a | CAA75080.1 | pET23a::hodC | [24] |
| AqdC | Mycobacteroides abscessus ATCC 19977 | CAM60402.1 | pET28b::his8-TEV-aqdCI | [17] |
| AqdCV | Mycobacteroides abscessus ATCC 19977 | CAM60402.1 | pBAD::his8-tev-aqdC- G40K-A134L-G220D-Y238W | [19] |
| AqdCVIII | Mycobacteroides abscessus ATCC 19977 | CAM60402.1 | pBAD::his8-tev-aqdC- G40K-G220D-Y238W | [19] |
| HQDN.f. | Nocardia farcinica IFM 10152 | BAD60071.1 | pET28b::his8-TEV- HQDN.f. | [14] |
| HQDS.b. | Streptomyces bingchenggensis BCW-1 | ADI11806.1 | pET28b::his8-TEV- HQDS.b. | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arranz San Martín, A.; Vogel, J.; Wullich, S.C.; Quax, W.J.; Fetzner, S. Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS): A Comparison between Naturally Occurring and Engineered PQS-Cleaving Dioxygenases. Biomolecules 2022, 12, 170. https://doi.org/10.3390/biom12020170
Arranz San Martín A, Vogel J, Wullich SC, Quax WJ, Fetzner S. Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS): A Comparison between Naturally Occurring and Engineered PQS-Cleaving Dioxygenases. Biomolecules. 2022; 12(2):170. https://doi.org/10.3390/biom12020170
Chicago/Turabian StyleArranz San Martín, Alba, Jan Vogel, Sandra C. Wullich, Wim J. Quax, and Susanne Fetzner. 2022. "Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS): A Comparison between Naturally Occurring and Engineered PQS-Cleaving Dioxygenases" Biomolecules 12, no. 2: 170. https://doi.org/10.3390/biom12020170
APA StyleArranz San Martín, A., Vogel, J., Wullich, S. C., Quax, W. J., & Fetzner, S. (2022). Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS): A Comparison between Naturally Occurring and Engineered PQS-Cleaving Dioxygenases. Biomolecules, 12(2), 170. https://doi.org/10.3390/biom12020170