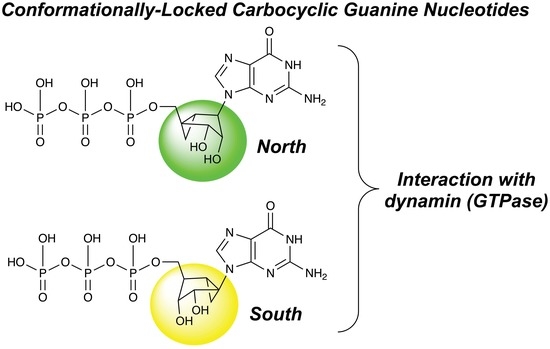
Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin
Abstract
:1. Introduction
2. Result and Discussion
2.1. Synthesis of Conformationally Locked Carbocyclic Guanine Nucleotides
2.2. Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin Activity
3. Experimental
3.1. Chemical Synthesis
3.2. Electron Microscopy of Dynamin-Mediated Constriction of Lipid Tubules
3.3. Computation of Puckering Parameters
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marquez, V.E.; Siddiqui, M.A.; Ezzitouni, A.; Russ, P.; Wang, J.; Wagner, R.W.; Matteucci, M.D. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J. Med. Chem. 1996, 39, 3739–3747. [Google Scholar] [CrossRef] [PubMed]
- Toti, K.S.; Osborne, D.; Ciancetta, A.; Boison, D.; Jacobson, K.A. South (S)- and North (N)-methanocarba-7-deazaadenosine analogues as inhibitors of human adenosine kinase. J. Med. Chem. 2016, 59, 6860–6877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, G.; Lee, K.; Ji, X.; Kim, H.S.; Soltysiak, K.A.; Marquez, V.E.; Jacobson, K.A. Synthesis and purine receptor affinity of 6-oxopurine nucleosides and nucleotides containing (N)-methanocarba-pseudoribose rings. Bioorg. Med. Chem. Lett. 2001, 11, 2295–2300. [Google Scholar] [CrossRef] [Green Version]
- Ravi, R.G.; Kim, H.S.; Servos, J.; Zimmermann, H.; Lee, K.; Maddileti, S.; Boyer, J.L.; Harden, T.K.; Jacobson, K.A. Adenine nucleotide analogues locked in a Northern methanocarba conformation: Enhanced stability and potency as P2Y1 receptor agonists. J. Med. Chem. 2002, 45, 2090–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinshaw, J.E. Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 2000, 16, 483–519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lee, D.M.; Jimah, J.R.; Gerassimov, N.; Yang, C.; Kim, S.; Luvsanjav, D.; Winkelman, J.; Mettlen, M.; Abrams, M.E.; et al. Dynamin regulates the dynamics and mechanical strength of the actin cytoskeleton as a multifilament actin-bundling protein. Nat. Cell Biol. 2002, 22, 674–688. [Google Scholar] [CrossRef]
- Kong, L.; Sochacki, K.A.; Wang, H.; Fang, S.; Canagarajah, B.; Kehr, A.D.; Rice, W.J.; Strub, M.-P.; Taraska, J.W.; Hinshaw, J.E. Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 2018, 560, 258–262. [Google Scholar] [CrossRef]
- Sundborger, A.C.; Fang, S.; Heymann, J.A.; Ray, P.; Chappie, J.S.; Hinshaw, J.E. A dynamin mutant defines a superconstricted prefission state. Cell Rep. 2014, 8, 734–742. [Google Scholar] [CrossRef] [Green Version]
- Jimah, J.R.; Hinshaw, J.E. Structural insights into the mechanism of dynamin superfamily proteins. Trends Cell Biol. 2019, 29, 257–273. [Google Scholar] [CrossRef]
- Melman, A.; Zhong, M.; Marquez, V.E.; Jacobson, K.A. Synthesis of enantiomerically pure (S)-methanocarbaribo uracil nucleoside derivatives for use as antiviral agents and P2Y receptor ligands. J. Org. Chem. 2008, 73, 8085–8088. [Google Scholar] [CrossRef] [Green Version]
- Dejmek, M.; Sála, M.; Plačková, P.; Hřebabecký, H.; Mascarell Borredà, L.; Neyts, J.; Dračínský, M.; Procházková, E.; Jansa, P.; Leyssen, P.; et al. Synthesis of novel purine-based coxsackievirus inhibitors bearing polycylic substituents at the N-9 position. Arch. Pharm. 2014, 347, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Caton-Williams, J.; Smith, M.; Carrasco, N.; Huang, Z. Protection-free one-pot synthesis of 2’-deoxynucleoside 5’-triphosphates and DNA polymerization. Org. Lett. 2011, 13, 4156–4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamady, S.; Desoky, A.; Taylor, S.D. Sulfonyl imidazolium salts as reagents for the rapid and efficient synthesis of nucleoside polyphosphates and their conjugates. Org. Lett. 2012, 14, 402–405. [Google Scholar] [CrossRef]
- Sweitzer, S.M.; Hinshaw, J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 1998, 93, 1021–1029. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Hinshaw, J.E. Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 2001, 3, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Calixto, A.R.; Moreira, C.; Kamerlin, S.C.L. Recent advances in understanding biological GTP hydrolysis through molecular simulation. ACS Omega 2020, 5, 4380–4385. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Jacobson, K.A. Survey of ribose ring pucker of signaling nucleosides and nucleotides. Nucleosides Nucleotides Nucleic. Acids 2020, 39, 322–341. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic. Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- RDKit: Open-Source Cheminformatics. Available online: http://www.rdkit.org (accessed on 15 March 2022).
- SMARTS Theory Manual, Daylight Chemical Information Systems, Santa Fe, New Mexico. Available online: https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html (accessed on 15 March 2022).
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]




| Gene and Protein Name (Species) | Uniprot Code | PDB Code | Ligand PDB Code (Ligand Description If Required) |
|---|---|---|---|
| DNM1, Dynamin-1 (Homo sapiens) | Q05193 | 2X2E 2X2F 3ZYC 5D3Q 6DLU 7AX3 | GDP GDP GCP (phosphomethylphosphonic acid guanylate ester) GDP GCP (phosphomethylphosphonic acid guanylate ester) GCP (phosphomethylphosphonic acid guanylate ester) |
| DNM1L, Dynamin-1-like protein (Homo sapiens) | O00429 | 3W6N 3W6O 3W6P 4H1V 5WP9 | GNH (aminophosphonic acid-guanylate ester) GCP (phosphomethylphosphonic acid guanylate ester) GDP GNP (phosphoaminophosphonic acid-guanylate ester) GCP (phosphomethylphosphonic acid guanylate ester) |
| DNM3, Dynamin-3 (Homo sapiens) | Q9UQ16 | 3L43 | GDP |
| OPA1, Dynamin-like 120 kDa protein, mitochondrial (Homo sapiens) | O60313 | 6JTG | GDP |
| MX1, Interferon-induced GTP-binding protein Mx1 (Homo sapiens) | P20591 | 4P4S 4P4T | GCP (phosphomethylphosphonic acid guanylate ester) GDP |
| MFN1, Mitofusin-1 (Homo sapiens) | Q8IWA4 | 5GNR 5GNS 5GNT 5GOE 5GOF 5GOM 5YEW | GDP GTP GDP GDP GTP GDP GDP |
| Ehd4, EH domain–containing protein 4 (Mus musculus) | Q9EQP2 | 5MTV 5MVF | AGS (phosphothiophosphoric acid-adenylate ester) ADP (adenosine-5′-diphosphate) |
| Npun_R6513, Bacterial dynamin-like protein (Nostoc punctiforme) | B2IZD3 | 2J68 2W6D | GDP GDP |
| DRP1A, Phragmoplastin DRP1A (Arabidopsis thaliana) | P42697 | 3T34 3T35 | GDP GDP |
| dymA, Dynamin-A (Dictyostelium discoideum) | Q94464 | 1JWY 1JX2 | GDP, ADP * (adenosine-5′-diphosphate) ADP * (adenosine-5′-diphosphate) * Not in dynamin binding site, but in myosin-fused domain |
| CTHT_0061810, Putative sorting protein (Chaetomium thermophilum) | G0SFF0 | 6DEF 6DI7 6DJQ 6VJF | GCP (phosphomethylphosphonic acid guanylate ester) GDP GDP GCP (phosphomethylphosphonic acid guanylate ester) |
| MGM1, Dynamin-like GTPase MGM1, mitochondrial (Saccharomyces cerevisiae) | P32266 | 6JSJ | GDP |
| Cj0412, Putative ATP/GTP binding protein (Campylobacter jejuni) | Q0PB98 | 5OXF | GDP |
| SEY1, Protein SEY1 (Candida albicans) | Q9C0L9 | 5CA8 5CA9 5CB2 | GDP GDP GNP (phosphoaminophosphonic acid-guanylate ester) |
| iniA, Isoniazid inducible gene protein IniA (Mycolicibacterium smegmatis) | I7FE16 | 6J72 | GTP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toti, K.S.; Jimah, J.R.; Salmaso, V.; Hinshaw, J.E.; Jacobson, K.A. Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin. Biomolecules 2022, 12, 584. https://doi.org/10.3390/biom12040584
Toti KS, Jimah JR, Salmaso V, Hinshaw JE, Jacobson KA. Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin. Biomolecules. 2022; 12(4):584. https://doi.org/10.3390/biom12040584
Chicago/Turabian StyleToti, Kiran S., John R. Jimah, Veronica Salmaso, Jenny E. Hinshaw, and Kenneth A. Jacobson. 2022. "Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin" Biomolecules 12, no. 4: 584. https://doi.org/10.3390/biom12040584
APA StyleToti, K. S., Jimah, J. R., Salmaso, V., Hinshaw, J. E., & Jacobson, K. A. (2022). Synthesis and Effect of Conformationally Locked Carbocyclic Guanine Nucleotides on Dynamin. Biomolecules, 12(4), 584. https://doi.org/10.3390/biom12040584