Dynamical Oligomerisation of Histidine Rich Intrinsically Disordered Proteins Is Regulated through Zinc-Histidine Interactions
Abstract
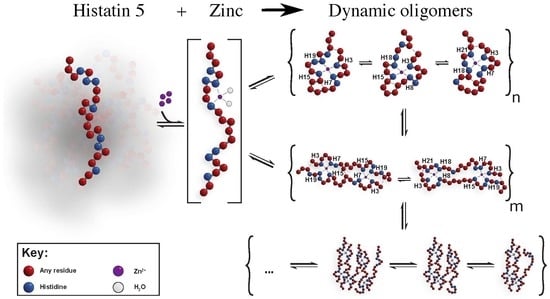
:1. Introduction
2. Materials And Methods
2.1. Sample Preparation
2.2. SAXS
2.3. NMR
2.4. Isothermal Titration Calorimetry
2.5. Coarse-Grained Model
2.6. Simulation Aspects
2.7. Structural Analysis
3. Results And Discussion
3.1. Tracing the Interaction Through Variants
3.2. Comparison with Computer Simulation
3.3. Stoichiometry, Coordination And Thermodynamics
4. Conclusions
Underlying Mechanism of The Oligomerisation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | linear dichroism |
References
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Ruff, K.M.; Pappu, R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015, 32, 102–112. [Google Scholar] [CrossRef]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Schad, E.; Tantos, A.; Kalmar, L. Intrinsically disordered proteins: Emerging interaction specialists. Curr. Opin. Struct. Biol. 2015, 35, 49–59. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef]
- Borg, M.; Mittag, T.; Pawson, T.; Tyers, M.; Forman-Kay, J.D.; Chan, H.S. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc. Natl. Acad. Sci. USA 2007, 104, 9650–9655. [Google Scholar] [CrossRef]
- Baran, C.; Smith, G.S.; Bamm, V.V.; Harauz, G.; Lee, J.S. Divalent cations induce a compaction of intrinsically disordered myelin basic protein. Biochem. Biophys. Res. Commun. 2010, 391, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Boys, B.L.; Brickenden, A.; Konermann, L.; Choy, W.Y. Effects of zinc binding on the structure and dynamics of the intrinsically disordered protein prothymosin α: Evidence for metalation as an entropic switch. Biochemistry 2007, 46, 13120–13130. [Google Scholar] [CrossRef]
- Faller, P.; Hureau, C.; La Penna, G. Metal Ions and Intrinsically Disordered Proteins and Peptides: From Cu/Zn Amyloid-β to General Principles. Accounts Chem. Res. 2014, 47, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Thorn, D.C.; Ecroyd, H.; Carver, J.A.; Holt, C. Casein structures in the context of unfolded proteins. Int. Dairy J. 2015, 46, 2–11. [Google Scholar] [CrossRef]
- Holt, C.; Lenton, S.; Nylander, T.; Sørensen, E.S.; Teixeira, S.C. Mineralisation of soft and hard tissues and the stability of biofluids. J. Struct. Biol. 2014, 185, 383–396. [Google Scholar] [CrossRef]
- Lenton, S.; Grimaldo, M.; Roosen-Runge, F.; Schreiber, F.; Nylander, T.; Clegg, R.; Holt, C.; Härtlein, M.; García Sakai, V.; Seydel, T.; et al. Effect of Phosphorylation on a Human-like Osteopontin Peptide. Biophys. J. 2017, 112, 1586–1596. [Google Scholar] [CrossRef]
- Lenton, S.; Seydel, T.; Nylander, T.; Holt, C.; Härtlein, M.; Teixeira, S.; Zaccai, G. Dynamic footprint of sequestration in the molecular fluctuations of osteopontin. J. R. Soc. Interface 2015, 12, 20150506. [Google Scholar] [CrossRef]
- Rieloff, E.; Tully, M.; Skepö, M. Assessing the Intricate Balance of Intermolecular Interactions upon Self-Association of Intrinsically Disordered Proteins. J. Mol. Biol. 2019, 431, 511–523. [Google Scholar] [CrossRef]
- Naganagowda, G.A.; Gururaja, T.L.; Levine, M.J. Delineation of conformational preferences in human salivary statherin by1H,31P NMR and CD Studies: Sequential Assignment and Structure-Function Correlations. J. Biomol. Struct. Dyn. 1998, 16, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Turoverov, K.K.; Kuznetsova, I.M.; Uversky, V.N. The protein kingdom extended: Ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog. Biophys. Mol. Biol. 2010, 102, 73–84. [Google Scholar] [CrossRef]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Proliferation of amyloid- 42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef]
- Istrate, A.N.; Kozin, S.A.; Zhokhov, S.S.; Mantsyzov, A.B.; Kechko, O.I.; Pastore, A.; Makarov, A.A.; Polshakov, V.I. Interplay of histidine residues of the Alzheimer’s disease Aβ peptide governs its Zn-induced oligomerization. Sci. Rep. 2016, 6, 21734. [Google Scholar] [CrossRef]
- Tõugu, V.; Karafin, A.; Palumaa, P. Binding of zinc(II) and copper(II) to the full-length Alzheimer’s amyloid-β peptide. J. Neurochem. 2008, 104, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Brewer, D.; Hunter, H.; Lajoie, G. NMR studies of the antimicrobial salivary peptides histatin 3 and histatin 5 in aqueous and nonaqueous solutions. Biochem. Cell Biol. 1998, 76, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Edgerton, M. How does it kill?: Understanding the candidacidal mechanism of salivary histatin 5. Eukaryot. Cell 2014, 13, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Raj, P.A.; Marcus, E.; Sukumaran, D.K. Structure of human salivary histatin 5 in aqueous and nonaqueous solutions. Biopolymers 1998, 45, 51–67. [Google Scholar] [CrossRef]
- Cragnell, C.; Durand, D.; Cabane, B.; Skepö, M. Coarse-grained modeling of the intrinsically disordered protein Histatin 5 in solution: Monte Carlo simulations in combination with SAXS. Proteins: Struct. Funct. Bioinform. 2016, 84, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Cragnell, C.; Rieloff, E.; Skepö, M. Utilizing Coarse-Grained Modeling and Monte Carlo Simulations to Evaluate the Conformational Ensemble of Intrinsically Disordered Proteins and Regions. J. Mol. Biol. 2018, 430, 2478–2492. [Google Scholar] [CrossRef] [PubMed]
- Situ, H.; Balasubramanian, S.V.; Bobek, L.A. Role of α-helical conformation of histatin-5 in candidacidal activity examined by proline variants. Biochim. Et Biophys. Acta Gen. Subj. 2000, 1475, 377–382. [Google Scholar] [CrossRef]
- Edgar, W.M.; Dawes, C.; O’Mullane, D.M. Saliva and Oral Health, 3rd ed.; British Dental Association: London, UK, 2004; p. 146. [Google Scholar]
- Conklin, S.E.; Bridgman, E.C.; Su, Q.; Riggs-Gelasco, P.; Haas, K.L.; Franz, K.J. Specific Histidine Residues Confer Histatin Peptides with Copper-Dependent Activity against Candida albicans. Biochemistry 2017, 56, 4244–4255. [Google Scholar]
- Grogan, J.; McKnight, C.J.; Troxler, R.F.; Oppenheim, F.G. Zinc and copper bind to unique sites of histatin 5. FEBS Lett. 2001, 491, 76–80. [Google Scholar] [CrossRef]
- Puri, S.; Li, R.; Ruszaj, D.; Tati, S.; Edgerton, M. Iron binding modulates candidacidal properties of salivary histatin 5. J. Dent. Res. 2015, 94, 201–208. [Google Scholar] [CrossRef]
- Ali, F.E.; Separovic, F.; Barrow, C.J.; Yao, S.; Barnham, K.J. Copper and Zinc Mediated Oligomerisation of Aβ Peptides. Int. J. Pept. Res. Ther. 2006, 12, 153–164. [Google Scholar] [CrossRef]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A. Computer programs ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef] [PubMed]
- Franke, D.; Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009, 42, 342–346. [Google Scholar] [PubMed]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef]
- Jephthah, S.; Staby, L.; Kragelund, B.B.; Skepö, M. Temperature Dependence of Intrinsically Disordered Proteins in Simulations: What are We Missing? J. Chem. Theory Comput. 2019, 15, 2672–2683. [Google Scholar] [CrossRef] [PubMed]
- Reščič, J.; Linse, P. MOLSIM: A modular molecular simulation software. J. Comput. Chem. 2015, 36, 1259–1274. [Google Scholar]
- Henriques, J.; Arleth, L.; Lindorff-Larsen, K.; Skepö, M. On the Calculation of SAXS Profiles of Folded and Intrinsically Disordered Proteins from Computer Simulations. J. Mol. Biol. 2018, 430, 2521–2539. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, M.; Kragelund, B.B.; Papaleo, E.; Lindorff-Larsen, K. An efficient method for estimating the hydrodynamic radius of disordered protein conformations. Biophys. J. 2017, 113, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Johansen, D.; Trewhella, J.; Goldenberg, D.P. Fractal dimension of an intrinsically disordered protein: Small-angle X-ray scattering and computational study of the bacteriophage λ N protein. Protein Sci. 2011, 20, 1955–1970. [Google Scholar] [CrossRef]
- Laitaoja, M.; Valjakka, J.; Jänis, J. Zinc Coordination Spheres in Protein Structures. Inorg. Chem. 2013, 52, 10983–10991. [Google Scholar] [CrossRef]
- Maret, W.; Li, Y. Coordination Dynamics of Zinc in Proteins. Chem. Rev. 2009, 109, 4682–4707. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, W.N.; Sträter, N. Recent Advances in Zinc Enzymology. Chem. Rev. 1996, 96, 2375–2434. [Google Scholar] [CrossRef] [PubMed]
- Minicozzi, V.; Stellato, F.; Comai, M.; Serra, M.D.; Potrich, C.; Meyer-Klaucke, W.; Morante, S. Identifying the Minimal Copper- and Zinc-binding Site Sequence in Amyloid-β Peptides. J. Biol. Chem. 2008, 283, 10784–10792. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrand, E.; Hermansson, K.; Friedman, R. Interaction Energies in Complexes of Zn and Amino Acids: A Comparison of Ab Initio and Force Field Based Calculations. J. Phys. Chem. A 2017, 121, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.M.; Kondrat, F.D.L.; Stewart, A.J.; Scrivens, J.H.; Sadler, P.J.; Blindauer, C.A. Native electrospray mass spectrometry approaches to probe the interaction between zinc and an anti-angiogenic peptide from histidine-rich glycoprotein. Sci. Rep. 2018, 8, 8646. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Miura, T.; Takeuchi, H. Inhibitory Effect of Copper(II) on Zinc(II)-Induced Aggregation of Amyloid β-Peptide. Biochem. Biophys. Res. Commun. 2001, 285, 991–996. [Google Scholar] [CrossRef] [PubMed]











| Kd (µM ) | N | ΔH (kJ/mol) | −TΔS (kJ/mol) | ΔG (kJ/mol) | |
|---|---|---|---|---|---|
| Hst5 | |||||
| Hst5DZM | |||||
| Hst5ZM |
| cp(mg/mL) | cp,Zn(mg/mL) | Rg(Å) | Rg,zn(Å) | I0 | I0,Zn | NH | ||
| Hst5 | 1.01 | 1.03 | 7 | 2.63 | ||||
| Hst5RAN | 0.94 | 0.94 | 7 | 1.96 | ||||
| Hst5DZM | 0.92 | 1.09 | 4 | 1.39 | ||||
| Hst5ZM | 1.13 | 1.23 | 3 | 1.48 | ||||
| Hst52H | 1.27 | 1.10 | 2 | 1.21 | ||||
| Hst5DH | 1.08 | 1.07 | 0 | 0.96 |
| NH | Rh (Å) | Rh,Zn (Å) | |
| Hst5 | 7 | 12.9 ± 0.7 | 12.9 ± 0.3 a |
| Hst5RAN | 7 | 12.9 ± 0.5 | 12.9 ± 0.3 a |
| Hst5DZM | 4 | 13.1 ± 0.4 | 14.2 ± 0.4 b |
| Hst5ZM | 3 | 13 ± 0.2 | 16.2 ± 0.9 b |
| Hst52H | 2 | 13 ± 0.2 | 13.8 ± 0.4 |
| Hst5DH | 0 | 12.6 ± 0.6 | 13.3 ± 0.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cragnell, C.; Staby, L.; Lenton, S.; Kragelund, B.B.; Skepö, M. Dynamical Oligomerisation of Histidine Rich Intrinsically Disordered Proteins Is Regulated through Zinc-Histidine Interactions. Biomolecules 2019, 9, 168. https://doi.org/10.3390/biom9050168
Cragnell C, Staby L, Lenton S, Kragelund BB, Skepö M. Dynamical Oligomerisation of Histidine Rich Intrinsically Disordered Proteins Is Regulated through Zinc-Histidine Interactions. Biomolecules. 2019; 9(5):168. https://doi.org/10.3390/biom9050168
Chicago/Turabian StyleCragnell, Carolina, Lasse Staby, Samuel Lenton, Birthe B. Kragelund, and Marie Skepö. 2019. "Dynamical Oligomerisation of Histidine Rich Intrinsically Disordered Proteins Is Regulated through Zinc-Histidine Interactions" Biomolecules 9, no. 5: 168. https://doi.org/10.3390/biom9050168
APA StyleCragnell, C., Staby, L., Lenton, S., Kragelund, B. B., & Skepö, M. (2019). Dynamical Oligomerisation of Histidine Rich Intrinsically Disordered Proteins Is Regulated through Zinc-Histidine Interactions. Biomolecules, 9(5), 168. https://doi.org/10.3390/biom9050168