Comparison of Independent Component Analysis, Principal Component Analysis, and Minimum Noise Fraction Transformation for Tree Species Classification Using APEX Hyperspectral Imagery
Abstract
:1. Introduction
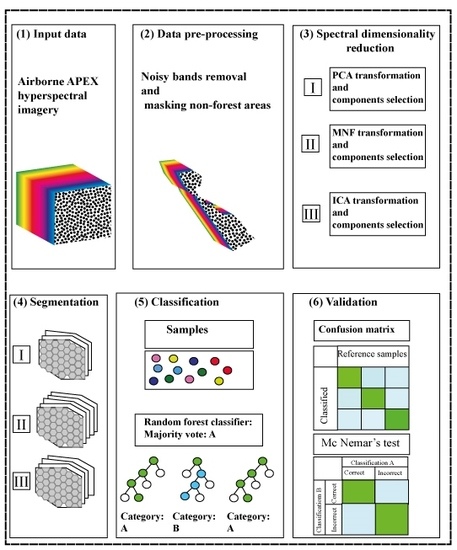
2. Materials and Methods
2.1. Study Area and Tree Species
2.2. Data
2.3. Methodology
2.4. Training and Validation Samples
2.5. Spectral Library
2.6. APEX Hyperspectral Image Preprocessing
2.7. Data Processing
2.8. Image Segmentation
2.9. Classification
2.10. Classification Accuracy Assessment
3. Results
3.1. Data Processing Results
3.2. Spectral Library of the Training Samples Using PCA, MNF, and ICA Inputs
3.3. Classification Results
3.4. Accuracy Assessment
3.5. Comparison of Classification Results According to McNemar’s Test
- Salix alba achieved a very good performance in all the classification results (producer’s accuracy of 98% to 100%).
- Populus x (hybrid): The best performance belonged to the ICA transformation. The poorest performance belonged to the results achieved from the APEX original image as an input (producer’s accuracy of 50%). The results achieved from the MNF and the PCA transformations were nearly the same (producer’s accuracy of nearly 95%).
- Picea abies also achieved a very good performance in all the classification results (producer’s accuracy of 98% to 100%).
- Alnus incana achieved a good classification result using DR data cubes as an input (producer’s accuracy of 65% to 100%). The poorest performance belonged to the original APEX hyperspectral image as an input (producer’s accuracy 86%).
- Fraxinus excelsior: The best performance of the classification belonged to the MNF, and ICA transformations and the original APEX hyperspectral imagery (producer’s accuracies of 95%, 93%, and 91%, respectively). The poorest performance belonged to the PCA transformation with a producer’s accuracy of 84%.
- Quercus robur had the poorest performance in comparison to the classification results of all other tree species. The best performance belonged to the ICA transformation (producer’s accuracy of 93%), and the poorest performance belonged to the original APEX as an input (producer’s accuracy of about 60%).
4. Discussion
4.1. Spectral Dimensionality Reduction
4.2. Segmentation and Classification
4.3. Tree Species Classification
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Reference | |||||||||
| Salix alba | Populus x (hybrid) | Picea abies | Alnus incana | Fraxinus excelsior | Quercus robur | Sum | Producer’s accuracy | ||
| Classified | Salix alba | 41 | 1 | 0 | 0 | 1 | 0 | 43 | 95% |
| Populus x (hybrid) | 0 | 22 | 4 | 14 | 3 | 43 | 50% | ||
| Picea abies | 0 | 0 | 42 | 0 | 1 | 0 | 43 | 98% | |
| Alnus incana | 0 | 1 | 0 | 37 | 0 | 5 | 43 | 86% | |
| Fraxinus excelsior | 0 | 0 | 0 | 3 | 39 | 1 | 43 | 91% | |
| Quercus robur | 1 | 1 | 2 | 6 | 6 | 27 | 43 | 63% | |
| Sum | 42 | 25 | 44 | 50 | 61 | 36 | 258 | ||
| User’s accuracy | 98% | 88% | 95% | 74% | 64% | 75% | |||
| Reference | |||||||||
| Salix alba | Populus x (hybrid) | Picea abies | Alnus incana | Fraxinus excelsior | Quercus robur | Sum | Producer’s accuracy | ||
| Classified | Salix alba | 41 | 1 | 1 | 0 | 0 | 0 | 43 | 95% |
| Populus x (hybrid) | 0 | 22 | 0 | 4 | 14 | 3 | 43 | 51% | |
| Picea abies | 0 | 0 | 42 | 0 | 1 | 43 | 98% | ||
| Alnus incana | 0 | 1 | 0 | 37 | 0 | 0 | 43 | 86% | |
| Fraxinus excelsior | 0 | 0 | 0 | 2 | 40 | 1 | 43 | 93% | |
| Quercus robur | 1 | 1 | 3 | 6 | 7 | 25 | 43 | 58% | |
| Sum | 42 | 25 | 46 | 49 | 62 | 29 | 258 | ||
| User’s accuracy | 98% | 88% | 91% | 76% | 65% | 74% | |||
| Reference | |||||||||
| Salix alba | Populus x (hybrid) | Picea abies | Alnus incana | Fraxinus excelsior | Quercus robur | Sum | Producer’s accuracy | ||
| Classified | Salix alba | 43 | 0 | 0 | 0 | 0 | 0 | 43 | 100% |
| Populus x (hybrid) | 0 | 40 | 1 | 2 | 0 | 43 | 93% | ||
| Picea abies | 0 | 0 | 42 | 0 | 1 | 43 | 98% | ||
| Alnus incana | 0 | 0 | 0 | 41 | 0 | 2 | 43 | 95% | |
| Fraxinus excelsior | 0 | 0 | 0 | 0 | 36 | 7 | 43 | 84% | |
| Quercus robur | 2 | 1 | 0 | 3 | 37 | 43 | 86% | ||
| Sum | 45 | 40 | 43 | 42 | 42 | 47 | 258 | ||
| User’s accuracy | 96% | 100% | 98% | 98% | 86% | 79% | |||
| Reference | |||||||||
| Salix alba | Populus x (hybrid) | Picea abies | Alnus incana | Fraxinus excelsior | Quercus robur | Sum | Producer’s accuracy | ||
| Classified | Salix alba | 43 | 0 | 0 | 0 | 0 | 0 | 43 | 100% |
| Populus x (hybrid) | 1 | 41 | 0 | 0 | 1 | 0 | 43 | 95% | |
| Picea abies | 0 | 0 | 43 | 0 | 0 | 0 | 43 | 100% | |
| Alnus incana | 0 | 0 | 0 | 41 | 2 | 0 | 43 | 95% | |
| Fraxinus excelsior | 0 | 0 | 0 | 0 | 41 | 2 | 43 | 95% | |
| Quercus robur | 2 | 0 | 0 | 1 | 4 | 36 | 43 | 84% | |
| Sum | 46 | 41 | 43 | 42 | 48 | 38 | 258 | ||
| User’s accuracy | 93% | 100% | 100% | 98% | 85% | 95% | |||
| Reference | |||||||||
| Salix alba | Populus x (hybrid) | Picea abies | Alnus incana | Fraxinus excelsior | Quercus robur | Sum | Producer’s accuracy | ||
| Classified | Salix alba | 43 | 0 | 0 | 0 | 0 | 0 | 43 | 100% |
| Populus x (hybrid) | 0 | 43 | 0 | 0 | 0 | 0 | 43 | 100% | |
| Picea abies | 0 | 0 | 43 | 0 | 0 | 0 | 43 | 100% | |
| Alnus incana | 0 | 0 | 0 | 43 | 0 | 0 | 43 | 100% | |
| Fraxinus excelsior | 0 | 0 | 0 | 0 | 40 | 3 | 43 | 93% | |
| Quercus robur | 0 | 0 | 0 | 0 | 3 | 40 | 43 | 93% | |
| Sum | 43 | 43 | 43 | 43 | 43 | 43 | 258 | ||
| User’s accuracy | 100% | 100% | 100% | 100% | 93% | 93% | |||
References
- Wetzel, F.T.; Hannu, S.; Eugenie, R.; Corinne, S.M.; Patricia, M.; Larissa, S.; Éamonn, Ó.T.; Francisco, A.G.C.; Anke, H.; Katrin, V. The roles and contributions of Biodiversity Observation Networks (BONs) in better tracking progress to 2020 biodiversity targets: A European case study. Biodiversity 2015, 16, 137–149. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, L.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Martin, M.E.; Newman, S.D.; Aber, J.D.; Congalton, R.G. Determining forest species composition using high spectral resolution remote sensing data. Remote Sen. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Sánchez-Azofeifa, A.; Castro-Esau, K. Intra-and inter-class spectral variability of tropical tree species at La Selva, Costa Rica: Implications for species identification using HYDICE imagery. Remote Sens. Environ. 2006, 105, 129–141. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Dalponte, M.; Orka, H.O.; Gobakken, T.; Gianelle, D.; Næsset, N. Tree species classification in boreal forests with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 101–110. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Becker, B.; Lusch, D.P.; Qi, J. A classification-based segmentation of the optimal spectral and spatial resolutions for Great Lakes coastal wetland imagery. Remote Sens. Environ. 2006, 108, 111–120. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.; Hughes, R.F. Invasive species detection in Hawaiian rainforests using airborne imaging spectroscopy and LiDAR. Remote Sens. Environ. 2008, 112, 1942–1955. [Google Scholar] [CrossRef]
- Oldeland, J.; Dorigo, W.; Wesuls, D.; Jürgens, N. Mapping bush encroaching species by seasonal differences in hyperspectral imagery. Remote Sens. 2010, 2, 1416–1438. [Google Scholar] [CrossRef]
- Harrison, D.; Rivard, B.; Sanchez-Azofeifa, A. Classification of tree species based on longwave hyperspectral data from leaves, a case study for a tropical dry forest. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 93–105. [Google Scholar] [CrossRef]
- Castro, K.L.; Sanchez-Azofeifa, G.A. Changes in spectral properties, chlorophyll content and internal mesophyll structure of senescing Populus balsamifera and Populus tremuloides leaves. Sensors 2008, 8, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Azofeifa, G.A.; Castro, K.; Wright, S.J.; Gamon, J.; Kalacska, M.; Rivard, B.; Schnitzer, S.A.; Feng, J.L. Differences in leaf traits, leaf internal structure, and spectral reflectance between two communities of lianas and trees: Implications for remote sensing in tropical environments. Remote Sens. Environ. 2009, 113, 2076–2088. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. Trans. Inform. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Richter, R.; Reu, B.; Wirth, C.; Doktor, D.; Vohland, M. The use of airborne hyperspectral data for tree species classification in a species-rich Central European forest area. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 464–474. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, J.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, 110–122. [Google Scholar] [CrossRef]
- Hamada, Y.; Stow, D.A.; Coulter, L.L.; Jafolla, J.C.; Hendricks, L.W. Detecting Tamarisk species (Tamarix spp.) in riparian habitats of Southern California using high spatial resolution hyperspectral imagery. Remote Sens. Environ. 2007, 109, 237–248. [Google Scholar] [CrossRef]
- Baatz, M.; Schäpe, A. Multiresolution segmentation-an optimization approach for high quality multi-scale image segmentation. In Angewandte Geographische Informationsverarbeitung; Strobl, J., Blaswchke, T., Griesebner, G., Eds.; Wischmann-Verlag: Heidelberg, Germany, 2000; Volume 12, pp. 12–23. [Google Scholar]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Blaschke, T.; Hay, G.J.; Kelly, M.; Lang, S.; Hofmann, P.; Addink, E.; Feitosa, R.Q.; Van der Meer, F.; Van der Werff, H.; Van Coillie, F.; et al. Geographic object-based image analysis—Towards a new paradigm. ISPRS J. Photogramm. Remote Sens. 2014, 87, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Corbane, C.; Pernkopf, L. Earth observation for habitat and biodiversity monitoring. In Creating the GIScociety, GI_Forum 2013, Salzburg, Austria, 2–5 July 2013; Wichmann-Verlag: Berlin, Germany, 2013; pp. 478–486. ISBN 978-3-87907-532-4. [Google Scholar] [CrossRef]
- Lang, S. Object-based image analysis for remote sensing applications: Modeling reality—Dealing with complexity. In Object Based Image Analysis-Spatial Concepts for Knowledge-Driven Remote Sensing Applications; Blaschke, T., Lang, S., Hay, G., Eds.; Springer: Berlin, Germany, 2008; pp. 3–28. ISBN 978-3-540-77057-2. [Google Scholar]
- Kamal, M.; Stuart, P. Hyperspectral data for mangrove species mapping: A comparison of pixel-based and object-based approach. Remote Sens. 2011, 3, 2222–2242. [Google Scholar] [CrossRef]
- Chen, G.; Weng, Q.; Hay, G.J.; He, Y. Geographic Object-based Image Analysis (GEOBIA): Emerging trends and future opportunities. GISci. Remote Sens. 2018, 55, 159–182. [Google Scholar] [CrossRef]
- Plaza, A.; Martinez, P.; Plaza, J.; Perez, R. Dimensionality reduction and classification of hyperspectral image data using sequences of extended morphological transformations. IEEE Trans. Geosci. Remote Sens. 2005, 43, 466–479. [Google Scholar] [CrossRef] [Green Version]
- Thenkabail, A.; Lyon, P.S.; Huete, J.G.; Bajwa, S.G.; Kulkarni, S.S. Hyperspectral Data Mining. In Hyperspectral Remote Sensing of Vegetation, 1st ed.; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; Chapter 4; pp. 93–120. [Google Scholar]
- Bajwa, S.G.; Bajcsy, P.; Groves, P.; Tian, L.F. Hyperspectral image data mining for band selection in agricultural applications. Trans. ASAE 2004, 47, 895–907. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advances in Hyperspectral Remote Sensing of Vegetation and Agricultural Croplands. In Hyperspectral Remote Sensing of Vegetation, 1st ed.; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; Chapter 1; pp. 3–35. [Google Scholar]
- Jolliffe, I. Principal component analysis. In International Encyclopedia of Statistical Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1094–1096. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Comon, P. Independent component analysis, a new concept? Signal Process. 1994, 36, 287–314. [Google Scholar] [CrossRef]
- Harsanyi, J.C.; Chang, C.I. Hyperspectral image classification and dimensionality reduction: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote Sens. 1994, 32, 779–785. [Google Scholar] [CrossRef]
- Underwood, E.; Ustin, S.; DiPietro, D. Mapping nonnative plants using hyperspectral imagery. Remote Sens. Environ. 2003, 86, 150–161. [Google Scholar] [CrossRef]
- Nascimento, J.M.P.; Dias, J.M.B. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Rodarmel, C.; Shan, J. Principal component analysis for hyperspectral image classification. Surv. Land Inf. Sci. 2002, 62, 115–122. [Google Scholar]
- Hyvarinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999, 10, 626–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, T.; Lang, S. Object-based class modelling for multi-scale riparian forest habitat mapping. Int. J. Appl. Earth Obs. Geoinf. 2014, 37, 29–37. [Google Scholar] [CrossRef]
- Biesemans, J.; Sterckx, S.; Knaeps, E.; Vreys, K.; Adriaensen, S.; Hooyberghs, J.; Meuleman, K.; Kempeneers, P.; Deronde, B.; Everaerts, J. Image processing workflows for airborne remote sensing. Paper Presented at the 5th EARSeL Workshop on Imaging Spectroscopy, Bruges, Belgium, 23–25 April 2007. [Google Scholar]
- Haan, J.F.; Kokke, J.M.M. Remote Sensing Algorithm Development: Toolkit I: Operationalization of Atmospheric Correction Methods for Tidal and Inland Waters; Remote Sensing Board (BCRS): Delft, The Netherlands, 1996; ISBN 9054112042. [Google Scholar]
- Jensen, R.J. Introductory Digital Image Processing, 3rd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2005. [Google Scholar]
- Cochrane, M.A. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2000, 21, 2075–2087. [Google Scholar] [CrossRef]
- Smith, G.; Curran, P. The signal-to-noise ratio (SNR) required for the estimation of foliar biochemical concentrations. Int. J. Remote Sens. 1996, 17, 1031–1058. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Zhang, Y.J. A survey on evaluation methods for image segmentation. Pattern Recognit. 1996, 29, 1335–1346. [Google Scholar] [CrossRef]
- Pal, N.R.; Pal, S.K. A review on image segmentation techniques. Pattern Recognit. 1993, 26, 1277–1294. [Google Scholar] [CrossRef]
- Haralick, R.M.; Linda, G.; Shapiro, L.G. Image segmentation techniques. Presented at the 1985 Technical Symposium East, Arlington, VA, USA, 8–12 April 1985. [Google Scholar]
- Liu, D.; Xia, F. Assessing object-based classification: Advantages and limitations. Remote Sens. Lett. 2010, 1, 187–194. [Google Scholar] [CrossRef]
- Belgiu, M.; Drǎguţ, D.; Strobl, J. Quantitative evaluation of variations in rule-based classifications of land cover in urban neighbourhoods using WorldView-2 imagery. ISPRS J. Photogramm. Remote Sens. 2014, 87, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Burnett, C.; Blaschke, T. A multi-scale segmentation/object relationship modelling methodology for landscape analysis. Ecol. Model. 2003, 168, 233–249. [Google Scholar] [CrossRef]
- Hay, G.J.; Castilla, G. Geographic object-based image analysis (geobia): A new name for a new discipline. In Object-Based Image Analysis; Blaschke, T., Lang, S., Hay, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 75–89. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC super-pixels compared to state-of-the-art super-pixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Csillik, O. Fast segmentation and classification of very high resolution remote sensing data using SLIC super-pixels. Remote Sens. 2017, 9, 243. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef]
- Liu, T.; Abd-Elrahman, A.; Morton, J.; Wilhelm, V.L. Comparing fully convolutional networks, random forest, support vector machine, and patch-based deep convolutional neural networks for object-based wetland mapping using images from small unmanned aircraft system. GISci. Remote Sens. 2018, 55, 243–264. [Google Scholar] [CrossRef]
- Guidici, D.; Clark, M.L. One-Dimensional convolutional neural network land-cover classification of multi-seasonal hyperspectral imagery in the San Francisco Bay Area, California. Remote Sens. 2017, 9, 629. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–spatial feature extraction for hyperspectral image classification: A dimension reduction and deep learning approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C.; Beard, k. H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Bosch, A.; Zisserman, A.; Munoz, X. Image classification using random forests and ferns. In Proceedings of the 2007 IEEE 11th International Conference on Computer Vision, Rio de Janerio, Brazil, 14–21 October 2007. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS. J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data: Principles and Practices, 2nd ed.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2009; ISBN 978-1-4200-5512-2. [Google Scholar]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Bradley, J.V. Distribution-Free Statistical Tests; Air Force Aerospace Medical Research Lab: Wright-Patterson AFB, OH, USA; Unwin Hyman: London, UK, 1968. [Google Scholar]
- Foody, G.M. Thematic map comparison. Photogramm. Eng. Remote Sens. 2004, 70, 627–633. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral dimensionality reduction for biophysical variable statistical retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Benz, U.C.; Hofmann, P.; Willhauck, G.; Lingenfelder, I.; Heynen, M. Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS J. Photogramm. Remote Sens. 2004, 58, 239–258. [Google Scholar] [CrossRef] [Green Version]
- Shah, C.A.; Arora, M.K.; Varshney, P.K. Unsupervised classification of hyperspectral data: An ICA mixture model based approach. Int. J. Remote Sens. 2004, 25, 481–487. [Google Scholar] [CrossRef]
- Blaschke, T.; Piralilou, S.T. The near-decomposability paradigm re-interpreted for place-based GIS. Presented at the 1st Workshop on Platial Analysis (PLATIAL’18), Heidelberg, Germany, 20–21 September 2018. [Google Scholar]

















| Bands | No. of Segments after Super-Pixel Segmentation | ||
|---|---|---|---|
| APEX image | 268 | 211,665 |  |
| PCA | 20 | 207,547 |  |
| MNF | 35 | 199,762 |  |
| ICA | 27 | 211,665 |  |
| ICA | MNF | PCA | APEX (Mtry = 268) | APEX (Mtry = 17) | |
|---|---|---|---|---|---|
| Overall accuracy (%) | 97 | 94 | 92 | 80 | 80 |
| Kappa coefficient | 0.972 | 0.939 | 0.911 | 0.767 | 0.762 |
| X2 | P | |
|---|---|---|
| ICA–MNF | 5.143 | 0.0233 |
| MNF–PCA | 1.25 | 0.2636 |
| ICA–PCA | 1.25 | 0.2636 |
| APEX (Mtry = 268)–APEX (Mtry = 17) | 1 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabiri, Z.; Lang, S. Comparison of Independent Component Analysis, Principal Component Analysis, and Minimum Noise Fraction Transformation for Tree Species Classification Using APEX Hyperspectral Imagery. ISPRS Int. J. Geo-Inf. 2018, 7, 488. https://doi.org/10.3390/ijgi7120488
Dabiri Z, Lang S. Comparison of Independent Component Analysis, Principal Component Analysis, and Minimum Noise Fraction Transformation for Tree Species Classification Using APEX Hyperspectral Imagery. ISPRS International Journal of Geo-Information. 2018; 7(12):488. https://doi.org/10.3390/ijgi7120488
Chicago/Turabian StyleDabiri, Zahra, and Stefan Lang. 2018. "Comparison of Independent Component Analysis, Principal Component Analysis, and Minimum Noise Fraction Transformation for Tree Species Classification Using APEX Hyperspectral Imagery" ISPRS International Journal of Geo-Information 7, no. 12: 488. https://doi.org/10.3390/ijgi7120488
APA StyleDabiri, Z., & Lang, S. (2018). Comparison of Independent Component Analysis, Principal Component Analysis, and Minimum Noise Fraction Transformation for Tree Species Classification Using APEX Hyperspectral Imagery. ISPRS International Journal of Geo-Information, 7(12), 488. https://doi.org/10.3390/ijgi7120488