Emerging Roles of DYRK Kinases in Embryogenesis and Hedgehog Pathway Control
Abstract
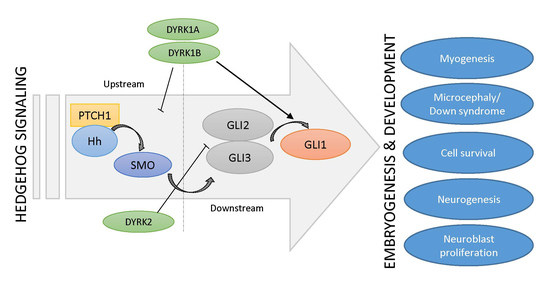
:1. Introduction
1.1. The Hedgehog Signaling Pathway
1.2. Protein Kinases: An Introduction
2. The CMGC Group of Kinases
3. The DYRK Family of Kinases
4. Class I DYRKs: DYRK1A and DYRK1B
4.1. The DYRK1A Kinase
4.2. DYRK1A as a Regulator of (Neuronal) Hedgehog Signaling
4.3. The DYRK1B Kinase
4.4. DYRK1B in Developmental and Physiological Processes
5. The Class II DYRKs
5.1. The DYRK2, DYRK3, and DYRK4 Kinases
5.2. Class II DYRKs in Development
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.T.; Zhao, Z.; Ingham, P.W. Hedgehog signalling. Development 2016, 143, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Di Magno, L.; Coni, S.; Di Marcotullio, L.; Canettieri, G. Digging a hole under Hedgehog: Downstream inhibition as an emerging anticancer strategy. Biochim. Biophys. Acta 2015, 1856, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Riddle, R.D.; Johnson, R.L.; Laufer, E.; Tabin, C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 1993, 75, 1401–1416. [Google Scholar] [CrossRef]
- Echelard, Y.; Epstein, D.J.; St-Jacques, B.; Shen, L.; Mohler, J.; McMahon, J.A.; McMahon, A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 1993, 75, 1417–1430. [Google Scholar] [CrossRef]
- Roelink, H.; Augsburger, A.; Heemskerk, J.; Korzh, V.; Norlin, S.; Ruiz i Altaba, A.; Tanabe, Y.; Placzek, M.; Edlund, T.; Jessell, T.M.; et al. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 1994, 76, 761–775. [Google Scholar] [CrossRef]
- Cohn, M.J.; Tickle, C. Limbs: A model for pattern formation within the vertebrate body plan. Trends Genet. 1996, 12, 253–257. [Google Scholar] [CrossRef]
- Jessell, T.M. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 2000, 1, 20–29. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.P.; Ingham, P.W.; Tabin, C.J. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 2003, 53, 1–114. [Google Scholar] [PubMed]
- St-Jacques, B.; Hammerschmidt, M.; McMahon, A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999, 13, 2072–2086. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Tong, M.; Jameson, J.L. Distinct roles for steroidogenic factor 1 and desert hedgehog pathways in fetal and adult Leydig cell development. Endocrinology 2007, 148, 3704–3710. [Google Scholar] [CrossRef] [PubMed]
- Pandit, T.; Ogden, S. Contributions of Noncanonical Smoothened Signaling During Embryonic Development. J. Dev. Biol. 2017, 5, 11. [Google Scholar] [CrossRef]
- Belgacem, Y.H.; Hamilton, A.M.; Shim, S.; Spencer, K.A.; Borodinsky, L.N. The Many Hats of Sonic Hedgehog Signaling in Nervous System Development and Disease. J. Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef]
- Ramsbottom, S.A.; Pownall, M.E. Regulation of Hedgehog Signalling Inside and Outside the Cell. J. Dev. Biol. 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Silva, H.; Correia-Pinto, J.; Moura, R. Canonical Sonic Hedgehog Signaling in Early Lung Development. J. Dev. Biol. 2017, 5, 3. [Google Scholar] [CrossRef]
- Nakano, Y.; Guerrero, I.; Hidalgo, A.; Taylor, A.; Whittle, J.R.S.; Ingham, P.W. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature 1989, 341, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E.; Scott, M.P. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell 1989, 59, 751–765. [Google Scholar] [CrossRef]
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature 2002, 418, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.; Casillas, C.; Alfaro, A.C.; Jagers, C.; Roelink, H. Patched1 and Patched2 inhibit Smoothened non-cell autonomously. Elife 2016, 5, e17634. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.C.; Angers, S. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 513–537. [Google Scholar] [CrossRef] [PubMed]
- Riobo, N.A.; Lu, K.; Ai, X.; Haines, G.M.; Emerson, C.P., Jr. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 4505–4510. [Google Scholar] [CrossRef] [PubMed]
- Lauth, M.; Bergstrom, A.; Toftgard, R. Phorbol esters inhibit the Hedgehog signalling pathway downstream of Suppressor of Fused, but upstream of Gli. Oncogene 2007, 26, 5163–5168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, Q.; Yen, C.J.; Xia, W.; Izzo, J.G.; Lang, J.Y.; Li, C.W.; Hsu, J.L.; Miller, S.A.; Wang, X.; et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012, 21, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Fallon, J.F.; Beachy, P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 2000, 100, 423–434. [Google Scholar] [CrossRef]
- Chen, Y.; Yue, S.; Xie, L.; Pu, X.H.; Jin, T.; Cheng, S.Y. Dual Phosphorylation of suppressor of fused (Sufu) by PKA and GSK3beta regulates its stability and localization in the primary cilium. J. Biol. Chem. 2011, 286, 13502–13511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Lee, R.T.; Pusapati, G.V.; Iyu, A.; Rohatgi, R.; Ingham, P.W. An essential role for Grk2 in Hedgehog signalling downstream of Smoothened. EMBO Rep. 2016, 17, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Dhanyamraju, P.K.; Lauth, M. DYRK1B blocks canonical and promotes non-canonical Hedgehog signaling through activation of the mTOR/AKT pathway. Oncotarget 2017, 8, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Lauth, M.; Bergstrom, A.; Shimokawa, T.; Tostar, U.; Jin, Q.; Fendrich, V.; Guerra, C.; Barbacid, M.; Toftgard, R. DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat. Struct. Mol. Biol. 2010, 17, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, T.; Tostar, U.; Lauth, M.; Palaniswamy, R.; Kasper, M.; Toftgard, R.; Zaphiropoulos, P.G. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J. Biol. Chem. 2008, 283, 14345–14354. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Maye, P.; Kogerman, P.; Tejedor, F.J.; Toftgard, R.; Xie, W.; Wu, G.; Wu, D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 2002, 277, 35156–35161. [Google Scholar] [CrossRef] [PubMed]
- Varjosalo, M.; Bjorklund, M.; Cheng, F.; Syvanen, H.; Kivioja, T.; Kilpinen, S.; Sun, Z.; Kallioniemi, O.; Stunnenberg, H.G.; He, W.W.; et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 2008, 133, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Gruber, W.; Hutzinger, M.; Elmer, D.P.; Parigger, T.; Sternberg, C.; Cegielkowski, L.; Zaja, M.; Leban, J.; Michel, S.; Hamm, S.; et al. DYRK1B as therapeutic target in Hedgehog/GLI-dependent cancer cells with Smoothened inhibitor resistance. Oncotarget 2016, 7, 7134–7148. [Google Scholar] [CrossRef] [PubMed]
- Keramati, A.R.; Fathzadeh, M.; Go, G.W.; Singh, R.; Choi, M.; Faramarzi, S.; Mane, S.; Kasaei, M.; Sarajzadeh-Fard, K.; Hwa, J.; et al. A form of the metabolic syndrome associated with mutations in DYRK1B. N. Engl. J. Med. 2014, 370, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Bayo-Fina, J.M.; Singh, R.; Kumar Dhanyamraju, P.; Holz, P.; Baier, A.; Fendrich, V.; Ramaswamy, A.; Baumeister, S.; Martinez, E.D.; et al. Identification of a novel actin-dependent signal transducing module allows for the targeted degradation of GLI1. Nat. Commun. 2015, 6, 8023. [Google Scholar] [CrossRef] [PubMed]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Lahiry, P.; Torkamani, A.; Schork, N.J.; Hegele, R.A. Kinase mutations in human disease: Interpreting genotype-phenotype relationships. Nat. Rev. Genet. 2010, 11, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Saavedra, D.; Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins Struct. Funct. Bioinform. 2007, 68, 893–914. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Plowman, G.D.; Hunter, T.; Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002, 27, 514–520. [Google Scholar] [CrossRef]
- Varjosalo, M.; Keskitalo, S.; Van Drogen, A.; Nurkkala, H.; Vichalkovski, A.; Aebersold, R.; Gstaiger, M. The protein interaction landscape of the human CMGC kinase grou. Cell Rep. 2013, 3, 1306–1320. [Google Scholar] [CrossRef] [PubMed]
- Garrett, S.; Broach, J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989, 3, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Soppa, U.; Becker, W. DYRK protein kinases. Curr. Biol. 2015, 25, R488–R489. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.; Weber, Y.; Wetzel, K.; Eirmbter, K.; Tejedor, F.J.; Joost, H.G. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 1998, 273, 25893–25902. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Laguna, A.; de la Luna, S. DYRK family of protein kinases: Evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011, 25, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Galceran, J.; de Graaf, K.; Tejedor, F.J.; Becker, W. The MNB/DYRK1A protein kinase: Genetic and biochemical properties. J. Neural. Transm. Suppl. 2003, 67, 139–148. [Google Scholar]
- Liu, Q.; Tang, Y.; Chen, L.; Liu, N.; Lang, F.; Liu, H.; Wang, P.; Sun, X. E3 Ligase SCFbetaTrCP-induced DYRK1A Protein Degradation Is Essential for Cell Cycle Progression in HEK293 Cells. J. Biol. Chem. 2016, 291, 26399–26409. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Song, W.J.; Chung, K.C. Function and regulation of Dyrk1A: Towards understanding Down syndrome. Cell Mol. Life Sci. 2009, 66, 3235–3240. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Estivill, X.; de la Luna, S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003, 116, 3099–3107. [Google Scholar] [CrossRef] [PubMed]
- Salichs, E.; Ledda, A.; Mularoni, L.; Alba, M.M.; de la Luna, S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009, 5, e1000397. [Google Scholar] [CrossRef] [PubMed]
- Lochhead, P.A.; Sibbet, G.; Morrice, N.; Cleghon, V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 2005, 121, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Kinstrie, R.; Luebbering, N.; Miranda-Saavedra, D.; Sibbet, G.; Han, J.; Lochhead, P.A.; Cleghon, V. Characterization of a domain that transiently converts class 2 DYRKs into intramolecular tyrosine kinases. Sci. Signal 2010, 3, ra16. [Google Scholar] [CrossRef] [PubMed]
- Walte, A.; Ruben, K.; Birner-Gruenberger, R.; Preisinger, C.; Bamberg-Lemper, S.; Hilz, N.; Bracher, F.; Becker, W. Mechanism of dual specificity kinase activity of DYRK1A. FEBS. J. 2013, 280, 4495–4511. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, M.; Roos, A.K.; Savitsky, P.; Filippakopoulos, P.; Kettenbach, A.N.; Olsen, J.V.; Gerber, S.A.; Eswaran, J.; Knapp, S.; Elkins, J.M. Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure 2013, 21, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Altafaj, X.; Aranda, S.; de la Luna, S. DYRK1A Autophosphorylation on Serine Residue 520 Modulates Its Kinase Activity via 14–3-3 Binding. Mol. Biol. Cell 2007, 18, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, K.; Herault, Y.; Lott, I.T.; Antonarakis, S.E.; Reeves, R.H.; Dierssen, M. Down syndrome: From understanding the neurobiology to therapy. J. Neurosci. 2010, 30, 14943–14945. [Google Scholar] [CrossRef] [PubMed]
- Presson, A.P.; Partyka, G.; Jensen, K.M.; Devine, O.J.; Rasmussen, S.A.; McCabe, L.L.; McCabe, E.R.B. Current Estimate of Down Syndrome Population Prevalence in the United States. J. Pediatr. 2013, 163, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Epstein, C.J. Down’s syndrome: Critical genes in a critical region. Nature 2006, 441, 582–583. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.C.; Fajardo, D.; Pena, A.; Sanchez, A.; Dominguez, M.C.; Satizabal, J.M.; Garcia-Vallejo, F. Global differential expression of genes located in the Down Syndrome Critical Region in normal human brain. Colomb. Med. (Cali) 2014, 45, 154–161. [Google Scholar] [PubMed]
- Korenberg, J.R.; Kawashima, H.; Pulst, S.M.; Ikeuchi, T.; Ogasawara, N.; Yamamoto, K.; Schonberg, S.A.; West, R.; Allen, L.; Magenis, E.; et al. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am. J. Hum. Genet. 1990, 47, 236–246. [Google Scholar] [PubMed]
- Tschop, K.; Conery, A.R.; Litovchick, L.; Decaprio, J.A.; Settleman, J.; Harlow, E.; Dyson, N. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011, 25, 814–830. [Google Scholar] [CrossRef] [PubMed]
- Litovchick, L.; Florens, L.A.; Swanson, S.K.; Washburn, M.P.; DeCaprio, J.A. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011, 25, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Tejedor, F.; Zhu, X.R.; Kaltenbach, E.; Ackermann, A.; Baumann, A.; Canal, I.; Heisenberg, M.; Fischbach, K.F.; Pongs, O. Minibrain: A new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 1995, 14, 287–301. [Google Scholar] [CrossRef]
- Contestabile, A.; Fila, T.; Ceccarelli, C.; Bonasoni, P.; Bonapace, L.; Santini, D.; Bartesaghi, R.; Ciani, E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus 2007, 17, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Thomazeau, A.; Lassalle, O.; Iafrati, J.; Souchet, B.; Guedj, F.; Janel, N.; Chavis, P.; Delabar, J.; Manzoni, O.J. Prefrontal Deficits in a Murine Model Overexpressing the Down Syndrome Candidate Gene Dyrk1a. J. Neurosci. 2014, 34, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Dierssen, M. Down syndrome: The brain in trisomic mode. Nat. Rev. Neurosci. 2012, 13, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Lagran, M.; Altafaj, X.; Gallego, X.; Marti, E.; Estivill, X.; Sahun, I.; Fillat, C.; Dierssen, M. Motor phenotypic alterations in TgDyrk1a transgenic mice implicate DYRK1A in Down syndrome motor dysfunction. Neurobiol. Dis. 2004, 15, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Van Bon, B.W.M.; Coe, B.P.; Bernier, R.; Green, C.; Gerdts, J.; Witherspoon, K.; Kleefstra, T.; Willemsen, M.H.; Kumar, R.; Bosco, P.; et al. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry 2016, 21, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Tejedor, F.J.; Hammerle, B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2010, 278, 223–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moller, R.S.; Kubart, S.; Hoeltzenbein, M.; Heye, B.; Vogel, I.; Hansen, C.P.; Menzel, C.; Ullmann, R.; Tommerup, N.; Ropers, H.H.; et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am. J. Hum. Genet. 2008, 82, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Duan, W.Y.; Yu, B.; Tong, D.L.; Cheng, C.; Zhang, Y.F.; Wu, W.; Ye, K.; Zhang, W.X.; Wu, M.; et al. Autism-associated Dyrk1a truncation mutants impair neuronal dendritic and spine growth and interfere with postnatal cortical development. Mol. Psychiatry 2017. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Lagran, M.; Benavides-Piccione, R.; Ballesteros-Yanez, I.; Calvo, M.; Morales, M.; Fillat, C.; Defelipe, J.; Ramakers, G.J.; Dierssen, M. Dyrk1A influences neuronal morphogenesis through regulation of cytoskeletal dynamics in mammalian cortical neurons. Cereb. Cortex 2012, 22, 2867–2877. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Sung, J.Y.; Park, J.; Song, W.J.; Chang, S.; Chung, K.C. Dyrk1A negatively regulates the actin cytoskeleton through threonine phosphorylation of N-WASP. J. Cell Sci. 2012, 125, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Sims, D.; Baum, B. Parallel RNAi screens across different cell lines identify generic and cell type-specific regulators of actin organization and cell morphology. Genome Biol. 2009, 10, R26. [Google Scholar] [CrossRef] [PubMed]
- Gockler, N.; Jofre, G.; Papadopoulos, C.; Soppa, U.; Tejedor, F.J.; Becker, W. Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS J. 2009, 276, 6324–6337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Park, J.; Song, W.J.; Chang, S. Overexpression of Dyrk1A causes the defects in synaptic vesicle endocytosis. Neurosignals 2010, 18, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Van Bon, B.W.M.; Hoischen, A.; Hehir-Kwa, J.; de Brouwer, A.P.M.; Ruivenkamp, C.; Gijsbers, A.C.J.; Marcelis, C.L.; de Leeuw, N.; Veltman, J.A.; Brunner, H.G.; et al. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin. Genet. 2011, 79, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Hogart, A.; Leung, K.N.; Wang, N.J.; Wu, D.J.; Driscoll, J.; Vallero, R.O.; Schanen, N.C.; LaSalle, J.M. Chromosome 15q11–13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J. Med. Genet. 2009, 46, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Strong, E.; Butcher, D.T.; Singhania, R.; Mervis, C.B.; Morris, C.A.; De Carvalho, D.; Weksberg, R.; Osborne, L.R. Symmetrical Dose-Dependent DNA-Methylation Profiles in Children with Deletion or Duplication of 7q11.23. Am. J. Hum. Genet. 2015, 97, 216–227. [Google Scholar] [CrossRef] [PubMed]
- F Ferent, J.; Traiffort, E. Hedgehog: Multiple Paths for Multiple Roles in Shaping the Brain and Spinal Cord. Neuroscientist 2015, 21, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Stecca, B.; Ruiz i Altaba, A. Brain as a paradigm of organ growth: Hedgehog-Gli signaling in neural stem cells and brain tumors. J. Neurobiol. 2005, 64, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Roessler, E.; Belloni, E.; Gaudenz, K.; Jay, P.; Berta, P.; Scherer, S.W.; Tsui, L.C.; Muenke, M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 1996, 14, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Traiffort, E.; Dubourg, C.; Faure, H.; Rognan, D.; Odent, S.; Durou, M.R.; David, V.; Ruat, M. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J. Biol. Chem. 2004, 279, 42889–42897. [Google Scholar] [CrossRef] [PubMed]
- Belloni, E.; Muenke, M.; Roessler, E.; Traverso, G.; Siegel-Bartelt, J.; Frumkin, A.; Mitchell, H.F.; Donis-Keller, H.; Helms, C.; Hing, A.V.; et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 1996, 14, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Shiohama, T.; Fujii, K.; Miyashita, T.; Mizuochi, H.; Uchikawa, H.; Shimojo, N. Brain morphology in children with nevoid basal cell carcinoma syndrome. Am. J. Med. Genet. A 2017, 173, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Roper, R.J.; Baxter, L.L.; Saran, N.G.; Klinedinst, D.K.; Beachy, P.A.; Reeves, R.H. Defective cerebellar response to mitogenic Hedgehog signaling in Down [corrected] syndrome mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Das, I.; Park, J.M.; Shin, J.H.; Jeon, S.K.; Lorenzi, H.; Linden, D.J.; Worley, P.F.; Reeves, R.H. Hedgehog agonist therapy corrects structural and cognitive deficits in a Down syndrome mouse model. Sci. Transl. Med. 2013, 5, 201ra120. [Google Scholar] [CrossRef] [PubMed]
- Dutka, T.; Hallberg, D.; Reeves, R.H. Chronic up-regulation of the SHH pathway normalizes some developmental effects of trisomy in Ts65Dn mice. Mech. Dev. 2015, 135, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Ehe, B.K.; Lamson, D.R.; Tarpley, M.; Onyenwoke, R.U.; Graves, L.M.; Williams, K.P. Identification of a DYRK1A-mediated phosphorylation site within the nuclear localization sequence of the hedgehog transcription factor GLI1. Biochem. Biophys. Res. Commun. 2017, 491, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Saade, M.; Gonzalez-Gobartt, E.; Escalona, R.; Usieto, S.; Marti, E. Shh-mediated centrosomal recruitment of PKA promotes symmetric proliferative neuroepithelial cell division. Nat. Cell Biol. 2017, 19, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wang, M.; Wang, J.; Huang, X.; Yang, R.; Gao, W.Q. Cell Division Mode Change Mediates the Regulation of Cerebellar Granule Neurogenesis Controlled by the Sonic Hedgehog Signaling. Stem. Cell Rep. 2015, 5, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Ferron, S.R.; Pozo, N.; Laguna, A.; Aranda, S.; Porlan, E.; Moreno, M.; Fillat, C.; de la Luna, S.; Sanchez, P.; Arbones, M.L.; et al. Regulated segregation of kinase Dyrk1A during asymmetric neural stem cell division is critical for EGFR-mediated biased signaling. Cell Stem Cell 2010, 7, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Hammerle, B.; Vera-Samper, E.; Speicher, S.; Arencibia, R.; Martinez, S.; Tejedor, F.J. Mnb/Dyrk1A is transiently expressed and asymmetrically segregated in neural progenitor cells at the transition to neurogenic divisions. Dev. Biol. 2002, 246, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Yin, X.; Gu, J.; Zhang, X.; Shi, J.; Qian, W.; Ji, Y.; Cao, M.; Gu, X.; Ding, F.; et al. Truncation and Activation of Dual Specificity Tyrosine Phosphorylation-regulated Kinase 1A by Calpain I: A molecular mechanism linked to tau pathology in alzheimer disease. J. Biol. Chem. 2015, 290, 15219–15237. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.M.; Ward, J.M.; Yew, C.C.; Kovochich, A.; Dawson, D.W.; Black, M.A.; Brett, B.T.; Sheetz, T.E.; Dupuy, A.J.; Chang, D.K.; et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. USA 2012, 109, 5934–5941. [Google Scholar] [CrossRef] [PubMed]
- Nizetic, D.; Groet, J. Tumorigenesis in Down’s syndrome: Big lessons from a small chromosome. Nat. Rev. Cancer 2012, 12, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Sussan, T.E.; Yang, A.; Li, F.; Ostrowski, M.C.; Reeves, R.H. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature 2008, 451, 73–75. [Google Scholar] [CrossRef] [PubMed]
- De Wit, N.J.; Burtscher, H.J.; Weidle, U.H.; Ruiter, D.J.; van Muijen, G.N. Differentially expressed genes identified in human melanoma cell lines with different metastatic behaviour using high density oligonucleotide arrays. Melanoma Res. 2002, 12, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Genovesi, L.A.; Ng, C.G.; Davis, M.J.; Remke, M.; Taylor, M.D.; Adams, D.J.; Rust, A.G.; Ward, J.M.; Ban, K.H.; Jenkins, N.A.; et al. Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups. Proc. Natl. Acad. Sci. USA 2013, 110, E4325–E4334. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.H.; Zaslavsky, A.; Lynch, R.C.; Britt, C.; Okada, Y.; Siarey, R.J.; Lensch, M.W.; Park, I.H.; Yoon, S.S.; Minami, T.; et al. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 2009, 459, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.E.; Watson, A.R.; Baker, M.; Jones, T.A.; D’Amico, G.; Robinson, S.D.; Joffre, C.; Garrido-Urbani, S.; Rodriguez-Manzaneque, J.C.; Martino-Echarri, E.; et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature 2010, 465, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Lo Celso, C.; Spencer-Dene, B.; Zouboulis, C.C.; Watt, F.M. HAN11 binds mDia1 and controls GLI1 transcriptional activity. J. Dermatol. Sci. 2006, 44, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Park, S.; Ewton, D.Z.; Friedman, E. The survival kinase Mirk/Dyrk1B is a downstream effector of oncogenic K-ras in pancreatic cancer. Cancer Res. 2007, 67, 7247–7255. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Sheppard, K.E.; Pearson, R.B.; Campbell, I.G.; Gorringe, K.L.; Simpson, K.J. Functional analysis of genes in regions commonly amplified in high-grade serous and endometrioid ovarian cancer. Clin. Cancer Res. 2013, 19, 1411–1421. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Deng, X.; Friedman, E. Mirk protein kinase is a mitogen-activated protein kinase substrate that mediates survival of colon cancer cells. Cancer Res. 2000, 60, 3631–3637. [Google Scholar] [PubMed]
- Becker, W.; Joost, H.G. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid. Res. Mol. Biol. 1999, 62, 1–17. [Google Scholar] [PubMed]
- Kentrup, H.; Becker, W.; Heukelbach, J.; Wilmes, A.; Schürmann, A.; Huppertz, C.; Kainulainen, H.; Joost, H.G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 1996, 271, 3488–3495. [Google Scholar] [CrossRef] [PubMed]
- Leder, S.; Weber, Y.; Altafaj, X.; Estivill, X.; Joost, H.G.; Becker, W. Cloning and characterization of DYRK1B, a novel member of the DYRK family of protein kinases. Biochem. Biophys. Res. Commun. 1999, 254, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Z.; Wang, C.Y.; Shi, J.D.; Ruan, Q.G.; Eckenrode, S.; Davoodi-Semiromi, A.; Kukar, T.; Gu, Y.; Lian, W.; Wu, D.; et al. Molecular cloning and characterization of the mouse and human TUSP gene, a novel member of the tubby superfamily. Gene 2001, 273, 275–284. [Google Scholar] [CrossRef]
- Gao, J.; Zheng, Z.; Rawal, B.; Schell, M.J.; Bepler, G.; Haura, E.B. Mirk/Dyrk1B, a novel therapeutic target, mediates cell survival in non-small cell lung cancer cells. Cancer Biol. Ther. 2009, 8, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Mercer, S.E.; Friedman, E. Mirk/Dyrk1B: A multifunctional dual-specificity kinase involved in growth arrest, differentiation, and cell survival. Cell Biochem. Biophys. 2006, 45, 303–315. [Google Scholar] [CrossRef]
- Deng, X.; Ewton, D.Z.; Pawlikowski, B.; Maimone, M.; Friedman, E. Mirk/dyrk1B is a Rho-induced kinase active in skeletal muscle differentiation. J. Biol. Chem. 2003, 278, 41347–41354. [Google Scholar] [CrossRef] [PubMed]
- Mercer, S.E.; Ewton, D.Z.; Deng, X.; Lim, S.; Mazur, T.R.; Friedman, E. Mirk/Dyrk1B mediates survival during the differentiation of C2C12 myoblasts. J. Biol. Chem. 2005, 280, 25788–25801. [Google Scholar] [CrossRef] [PubMed]
- Mercer, S.E.; Ewton, D.Z.; Shah, S.; Naqvi, A.; Friedman, E. Mirk/Dyrk1b mediates cell survival in rhabdomyosarcomas. Cancer Res. 2006, 66, 5143–5150. [Google Scholar] [CrossRef] [PubMed]
- Koleva, M.; Kappler, R.; Vogler, M.; Herwig, A.; Fulda, S.; Hahn, H. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol. Life Sci. 2005, 62, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.; Regl, G.; Hofbauer, S.W.; Altenhofer, P.; Achatz, G.; Dlugosz, A.; Schnidar, H.; Greil, R.; Hartmann, T.N.; Aberger, F. Hedgehog/GLI and PI3K signaling in the initiation and maintenance of chronic lymphocytic leukemia. Oncogene 2015, 34, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Meliton, V.; Kim, W.K.; Lee, K.B.; Wang, J.C.; Nguyen, K.; Yoo, D.; Jung, M.E.; Atti, E.; Tetradis, S.; et al. Novel oxysterols have pro-osteogenic and anti-adipogenic effects in vitro and induce spinal fusion in vivo. J. Cell Biochem. 2011, 112, 1673–1684. [Google Scholar] [CrossRef] [PubMed]
- Kha, H.T.; Basseri, B.; Shouhed, D.; Richardson, J.; Tetradis, S.; Hahn, T.J.; Parhami, F. Oxysterols regulate differentiation of mesenchymal stem cells: Pro-bone and anti-fat. J. Bone Miner. Res. 2004, 19, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Nosavanh, L.; Yu, D.H.; Jaehnig, E.J.; Tong, Q.; Shen, L.; Chen, M.H. Cell-autonomous activation of Hedgehog signaling inhibits brown adipose tissue development. Proc. Natl. Acad. Sci. USA 2015, 112, 5069–5074. [Google Scholar] [CrossRef] [PubMed]
- Abu Jhaisha, S.; Widowati, E.W.; Kii, I.; Sonamoto, R.; Knapp, S.; Papadopoulos, C.; Becker, W. DYRK1B mutations associated with metabolic syndrome impair the chaperone-dependent maturation of the kinase domain. Sci. Rep. 2017, 7, 6420. [Google Scholar] [CrossRef] [PubMed]
- Lord, K.A.; Creasy, C.L.; King, A.G.; King, C.; Burns, B.M.; Lee, J.C.; Dillon, S.B. REDK, a novel human regulatory erythroid kinase. Blood 2000, 95, 2838. [Google Scholar] [PubMed]
- Taira, N.; Nihira, K.; Yamaguchi, T.; Miki, Y.; Yoshida, K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. Cell 2007, 25, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Taira, N.; Yamamoto, H.; Yamaguchi, T.; Miki, Y.; Yoshida, K. ATM augments nuclear stabilization of DYRK2 by inhibiting MDM2 in the apoptotic response to DNA damage. J. Biol. Chem. 2010, 285, 4909–4919. [Google Scholar] [CrossRef] [PubMed]
- Taira, N.; Mimoto, R.; Kurata, M.; Yamaguchi, T.; Kitagawa, M.; Miki, Y.; Yoshida, K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J. Clin. Investig. 2012, 122, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, K.; Chen, S.; Sun, Q.; Zhang, Y.; Chen, L.; Sun, X. NFATc1 phosphorylation by DYRK1A increases its protein stability. PLoS ONE 2017, 12, e0172985. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, N.; Hirota, T.; Sakai, M.; Sanada, K.; Fukada, Y. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell Biol. 2010, 30, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Ewton, D.Z.; Deng, X.; Mercer, S.E.; Friedman, E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J. Biol. Chem. 2004, 279, 27790–27798. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Mercer, S.E.; Shah, S.; Ewton, D.Z.; Friedman, E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G by Mirk/dyrk1B kinase. J. Biol. Chem. 2004, 279, 22498–22504. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Mimoto, R.; Yanaihara, N.; Imawari, Y.; Hirooka, S.; Okamoto, A.; Yoshida, K. DYRK2 regulates epithelial-mesenchymal-transition and chemosensitivity through Snail degradation in ovarian serous adenocarcinoma. Tumour Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Maddika, S.; Chen, J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 2009, 11, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jiao, S.; Tan, X.; Zhang, P.; You, F. DYRK2 displays muscle fiber type specific function during zebrafish early somitogenesis. Int. J. Dev. Biol. 2017, 61, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Jackson, H.E.; Ingham, P.W. Control of muscle fibre-type diversity during embryonic development: The zebrafish paradigm. Mech. Dev. 2013. [Google Scholar] [CrossRef] [PubMed]
- Luebbering, N.; Charlton-Perkins, M.; Kumar, J.P.; Lochead, P.A.; Rollmann, S.M.; Cook, T.; Cleghon, V. Drosophila Dyrk2 plays a role in the development of the visual system. PLoS ONE 2013, 8, e76775. [Google Scholar] [CrossRef] [PubMed]
- Strutt, D.I.; Mlodzik, M. Hedgehog is an indirect regulator of morphogenetic furrow progression in the Drosophila eye disc. Development 1997, 124, 3233–3240. [Google Scholar] [PubMed]
- Baker, N.E.; Bhattacharya, A.; Firth, L.C. Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev. Biol. 2009, 335, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Bogacheva, O.; Bogachev, O.; Menon, M.; Dev, A.; Houde, E.; Valoret, E.I.; Prosser, H.M.; Creasy, C.L.; Pickering, S.J.; Grau, E.; et al. DYRK3 Dual-specificity Kinase Attenuates Erythropoiesis during Anemia. J. Biol. Chem. 2008, 283, 36665–36675. [Google Scholar] [CrossRef] [PubMed]
- Sacher, F.; Moller, C.; Bone, W.; Gottwald, U.; Fritsch, M. The expression of the testis-specific Dyrk4 kinase is highly restricted to step 8 spermatids but is not required for male fertility in mice. Mol. Cell Endocrinol. 2007, 267, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Cridland, S.O.; Keys, J.R.; Papathanasiou, P.; Perkins, A.C. Indian hedgehog supports definitive erythropoiesis. Blood Cells Mol. Dis. 2009, 43, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.I.; Outram, S.V.; Saldana, J.I.; Furmanski, A.L.; Dessens, J.T.; Crompton, T. Regulation of murine normal and stress-induced erythropoiesis by Desert Hedgehog. Blood 2012, 119, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Paulson, R.F.; Shi, L.; Wu, D.C. Stress erythropoiesis: New signals and new stress progenitor cells. Curr. Opin. Hematol. 2011, 18, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Pelkmans, L.; Fava, E.; Grabner, H.; Hannus, M.; Habermann, B.; Krausz, E.; Zerial, M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 2005, 436, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Tang, L.Y.; Tang, Y.; Tang, Y.; Shen, Q.H.; Ding, J.; Chen, Y.; Zhang, Z.; Yu, T.T.; Zhang, Y.E.; et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. Elife 2014, 3, e02555. [Google Scholar] [CrossRef] [PubMed]
- Karpen, H.E.; Bukowski, J.T.; Hughes, T.; Gratton, J.P.; Sessa, W.C.; Gailani, M.R. The sonic hedgehog receptor patched associates with caveolin-1 in cholesterol-rich microdomains of the plasma membrane. J. Biol. Chem. 2001, 276, 19503–19511. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Diehl, A.M.; Li, Y.X. Sonic hedgehog ligand partners with caveolin-1 for intracellular transport. Lab. Investig. 2009, 89, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Song, W.; Zhu, C.; Xu, W.; Liu, H.; Zhang, S.; Huifang, L. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sahin, Z.; Szczepny, A.; McLaughlin, E.A.; Meistrich, M.L.; Zhou, W.; Ustunel, I.; Loveland, K.L. Dynamic Hedgehog signalling pathway activity in germline stem cells. Andrology 2014, 2, 267–274. [Google Scholar] [CrossRef] [PubMed]
- O'Hara, W.A.; Azar, W.J.; Behringer, R.R.; Renfree, M.B.; Pask, A.J. Desert hedgehog is a mammal-specific gene expressed during testicular and ovarian development in a marsupial. BMC Dev. Biol. 2011, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.H.; Whoriskey, W.; Capel, B. Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002, 16, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Sharma, S.; Nardone, J.; Tanasa, B.; Iuga, A.; Srikanth, S.; Okamura, H.; Bolton, D.; Feske, S.; Hogan, P.G.; et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature 2006, 441, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Frattini, V.; Bansal, M.; Castano, A.M.; Sherman, D.; Hutchinson, K.; Bruce, J.N.; Califano, A.; Liu, G.; Cardozo, T.; et al. An ID2-dependent mechanism for VHL inactivation in cancer. Nature 2016, 529, 172–177. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.; Lauth, M. Emerging Roles of DYRK Kinases in Embryogenesis and Hedgehog Pathway Control. J. Dev. Biol. 2017, 5, 13. https://doi.org/10.3390/jdb5040013
Singh R, Lauth M. Emerging Roles of DYRK Kinases in Embryogenesis and Hedgehog Pathway Control. Journal of Developmental Biology. 2017; 5(4):13. https://doi.org/10.3390/jdb5040013
Chicago/Turabian StyleSingh, Rajeev, and Matthias Lauth. 2017. "Emerging Roles of DYRK Kinases in Embryogenesis and Hedgehog Pathway Control" Journal of Developmental Biology 5, no. 4: 13. https://doi.org/10.3390/jdb5040013
APA StyleSingh, R., & Lauth, M. (2017). Emerging Roles of DYRK Kinases in Embryogenesis and Hedgehog Pathway Control. Journal of Developmental Biology, 5(4), 13. https://doi.org/10.3390/jdb5040013