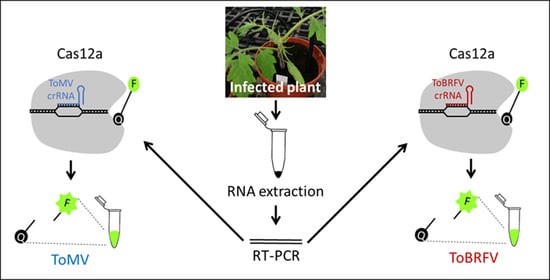
Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Virus Inoculation
2.2. RT-PCR Analysis
2.3. CRISPR RNA (crRNA) Design
2.4. In Vitro crRNA Transcription
2.5. LbCas12a Cleavage Assays
3. Results
3.1. Differential Detection of ToMV and ToBRFV Using CRISPR/Cas12a
3.2. Sensitivity and Specificity of CRISPR/Cas12a-Based Detection
3.3. CRISPR/Cas12a-Based Detection of ToBRFV from Field Samples
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber green mottle mosaic virus: Rapidly Increasing Global Distribution, Etiology, Epidemiology, and Management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Oladokun, J.O.; Halabi, M.H.; Barua, P.; Nath, P.D. Tomato brown rugose fruit disease: Current distribution, knowledge and future prospects. Plant Pathol. 2019, 68, 1579–1586. [Google Scholar] [CrossRef] [Green Version]
- Broadbent, L. Epidemiology and Control of Tomato Mosaic Virus. Annu. Rev. Phytopathol. 1976, 14, 75–96. [Google Scholar] [CrossRef]
- Dawson, W.O.; Beck, D.L.; Knorr, D.A.; Grantham, G.L. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc. Natl. Acad. Sci. USA 1986, 83, 1832–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goelet, P.; Lomonossoff, G.P.; Butler, P.J.; Akam, M.E.; Gait, M.J.; Karn, J. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 1982, 79, 5818–5822. [Google Scholar] [CrossRef] [Green Version]
- Reingold, V.; Lachman, O.; Belausov, E.; Koren, A.; Mor, N.; Dombrovsky, A. Epidemiological study ofCucumber green mottle mosaic virusin greenhouses enables reduction of disease damage in cucurbit production: Reducing CGMMV damage in greenhouses. Ann. Appl. Biol. 2016, 168, 29–40. [Google Scholar] [CrossRef]
- Tomlinson, J.A. Epidemiology and control of virus diseases of vegetables. Ann. Appl. Biol. 1987, 110, 661–681. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Barone, S.; Lo Bosco, G.; Rangel, E.A.; Davino, S. Spread of Tomato Brown Rugose Fruit Virus in Sicily and Evaluation of the Spatiotemporal Dispersion in Experimental Conditions. Agronomy 2020, 10, 834. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Smith, E. Seed Transmission of Tobamoviruses: Aspects of Global Disease Distribution. In Seed Biology; Jimenez-Lopez, J.C., Ed.; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar]
- Davino, S.; Caruso, A.G.; Bertacca, S.; Barone, S.; Panno, S. Tomato Brown Rugose Fruit Virus: Seed Transmission Rate and Efficacy of Different Seed Disinfection Treatments. Plants 2020, 9, 1615. [Google Scholar] [CrossRef]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A New Israeli Tobamovirus Isolate Infects Tomato Plants Harboring Tm-22 Resistance Genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [Green Version]
- Hak, H.; Spiegelman, Z. The Tomato brown rugose fruit virus movement protein overcomes Tm-22 resistance in tomato while attenuating viral transport. Mol. Plant. Microbe. Interact. 2021. [Google Scholar] [CrossRef]
- Beris, D.; Malandraki, I.; Kektsidou, O.; Theologidis, I.; Vassilakos, N.; Varveri, C. First Report of Tomato Brown Rugose Fruit Virus Infecting Tomato in Greece. Plant Dis. 2020, 104, 2035. [Google Scholar] [CrossRef] [Green Version]
- Alfaro-Fernández, A.; Castillo, P.; Sanahuja, E.; Rodríguez-Salido, M.D.C.; Font, M.I. First report of Tomato brown rugose fruit virus in tomato in Spain. Plant Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- van de Vossenberg, B.T.L.H.; Visser, M.; Bruinsma, M.; Koenraadt, H.M.S.; Westenberg, M.; Botermans, M. Real-time tracking of Tomato brown rugose fruit virus (ToBRFV) outbreaks in the Netherlands using Nextstrain. PLoS ONE 2020, 15, e0234671. [Google Scholar] [CrossRef] [PubMed]
- Skelton, A.; Buxton-Kirk, A.; Ward, R.; Harju, V.; Frew, L.; Fowkes, A.; Long, M.; Negus, A.; Forde, S.; Adams, I.P.; et al. First report of Tomato brown rugose fruit virus in tomato in the United Kingdom. New Dis. Rep. 2019, 40, 12. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Caruso, A.G.; Davino, S. First Report of Tomato Brown Rugose Fruit Virus on Tomato Crops in Italy. Plant Dis. 2019, 103, 1443. [Google Scholar] [CrossRef]
- Menzel, W.; Knierim, D.; Winter, S.; Hamacher, J.; Heupel, M. First report of tomato brown rugose fruit virus infecting tomato in Germany. New Dis. Rep. 2019, 39, 1. [Google Scholar] [CrossRef] [Green Version]
- Camacho-Beltrán, E.; Pérez-Villarreal, A.; Leyva-López, N.E.; Rodríguez-Negrete, E.A.; Ceniceros-Ojeda, E.A.; Méndez-Lozano, J. Occurrence of Tomato brown rugose fruit virus Infecting Tomato Crops in Mexico. Plant Dis. 2019, 103, 1440. [Google Scholar] [CrossRef]
- Ling, K.-S.; Tian, T.; Gurung, S.; Salati, R.; Gilliard, A. First Report of Tomato Brown Rugose Fruit Virus Infecting Greenhouse Tomato in the United States. Plant Dis. 2019, 103, 1439. [Google Scholar] [CrossRef]
- Yan, Z.-Y.; Ma, H.-Y.; Han, S.-L.; Geng, C.; Tian, Y.-P.; Li, X.-D. First Report of Tomato brown rugose fruit virus Infecting Tomato in China. Plant Dis. 2019, 103, 2973. [Google Scholar] [CrossRef]
- Fidan, H.; Sarikaya, P.; Calis, O. First report of Tomato brown rugose fruit virus on tomato in Turkey. New Dis. Rep. 2019, 39, 18. [Google Scholar] [CrossRef] [Green Version]
- Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants. Viruses 2020, 12, 879. [Google Scholar] [CrossRef] [PubMed]
- Klap, C.; Luria, N.; Smith, E.; Bakelman, E.; Belausov, E.; Laskar, O.; Lachman, O.; Gal-On, A.; Dombrovsky, A. The Potential Risk of Plant-Virus Disease Initiation by Infected Tomatoes. Plants 2020, 9, 623. [Google Scholar] [CrossRef]
- Levitzky, N.; Smith, E.; Lachman, O.; Luria, N.; Mizrahi, Y.; Bakelman, H.; Sela, N.; Laskar, O.; Milrot, E.; Dombrovsky, A. The bumblebee Bombus terrestris carries a primary inoculum of Tomato brown rugose fruit virus contributing to disease spread in tomatoes. PLoS ONE 2019, 14, e0210871. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Mendoza, J.; de Jesús García-Ávila, C.; López-Buenfil, J.A.; Araujo-Ruiz, K.; Quezada-Salinas, A.; Cambrón-Crisantos, J.M.; Ochoa-Martínez, D.L. Identificación de Tomato brown rugose fruit virus por RT-PCR de una región codificante de la replicasa (RdRP). Rev. Mex. Fitopatol. Mex. J. Phytopathol. 2019, 37. [Google Scholar] [CrossRef] [Green Version]
- Alkowni, R.; Alabdallah, O.; Fadda, Z. Molecular identification of tomato brown rugose fruit virus in tomato in Palestine. J. Plant Pathol. 2019, 101, 719–723. [Google Scholar] [CrossRef]
- Abstracts of Presentations at Plant Health 2019. Phytopathology 2019, 109, S2.1–S2.191.
- Rizzo, D.; Da Lio, D.; Panattoni, A.; Salemi, C.; Cappellini, G.; Bartolini, L.; Parrella, G. Rapid and Sensitive Detection of Tomato Brown Rugose Fruit Virus in Tomato and Pepper Seeds by Reverse Transcription Loop-Mediated Isothermal Amplification Assays (Real Time and Visual) and Comparison with RT-PCR End-Point and RT-qPCR Methods. Front. Microbiol. 2021, 12, 640932. [Google Scholar] [CrossRef]
- Sarkes, A.; Fu, H.; Feindel, D.; Harding, M.; Feng, J. Development and evaluation of a loop-mediated isothermal amplification (LAMP) assay for the detection of Tomato brown rugose fruit virus (ToBRFV). PLoS ONE 2020, 15, e0230403. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [Green Version]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [Green Version]
- Charpentier, E.; Marraffini, L.A. Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr. Opin. Microbiol. 2014, 19, 114–119. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garst, A.D.; Bassalo, M.C.; Pines, G.; Lynch, S.A.; Halweg-Edwards, A.L.; Liu, R.; Liang, L.; Wang, Z.; Zeitoun, R.; Alexander, W.G.; et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 2017, 35, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Jiao, J.; Kong, K.; Han, J.; Song, S.; Bai, T.; Song, C.; Wang, M.; Yan, Z.; Zhang, H.; Zhang, R.; et al. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 2021, 19, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Mahas, A.; Marsic, T.; Hassan, N.; Mahfouz, M.M. Efficient, Rapid, and Sensitive Detection of Plant RNA Viruses with One-Pot RT-RPA-CRISPR/Cas12a Assay. Front. Microbiol. 2020, 11, 610872. [Google Scholar] [CrossRef]
- Mahas, A.; Hassan, N.; Aman, R.; Marsic, T.; Wang, Q.; Ali, Z.; Mahfouz, M.M. LAMP-Coupled CRISPR–Cas12a Module for Rapid and Sensitive Detection of Plant DNA Viruses. Viruses 2021, 13, 466. [Google Scholar] [CrossRef] [PubMed]
- Tomato Mosaic Virus and Tobacco Mosaic Virus. Available online: https://extension.umn.edu/diseases/tomato-mosaic-virus-and-tobacco-mosaic-virus (accessed on 13 June 2021).
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Panno, S.; Ruiz-Ruiz, S.; Caruso, A.G.; Alfaro-Fernandez, A.; Font San Ambrosio, M.I.; Davino, S. Real-time reverse transcription polymerase chain reaction development for rapid detection of Tomato brown rugose fruit virus and comparison with other techniques. Peer J. 2019, 7, e7928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lim, J.; Shin, M.; Paek, S.-H.; Choi, J.-W. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2020. [Google Scholar] [CrossRef]
- Liu, P.-F.; Zhao, K.-R.; Liu, Z.-J.; Wang, L.; Ye, S.-Y.; Liang, G.-X. Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens. Bioelectron. 2021, 176, 112954. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2020. [Google Scholar] [CrossRef]




| Name (* nt Position) | Sequence (5′-3′) |
|---|---|
| T7-crRNA-F | ATCTACAACAGTAGAAATTCCCTATAGTGAGTCGTATTAATTTC |
| tobrfv-3 (3023–3049) | ** ACCGACGATGTACACATGACATGatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tobrfv-5 (2826–2852) | GCACAGAGACATAGAAACAAGAAatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tobrfv-9 (1822–1848) | GAGGTAGACCCAATGACTGCAGCatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tomv-1 (2469–2495) | GCCATATCAGAATATACTACCGAatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tomv-7 (2295–2321) | GAAGCAACATCCAGAACTCCGAAatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tomv-8 (1746–1772) | AGTACAGACAGTTCGGACAGTGCatctacaacagtagaaattccctatagtgagtcgtattaatttc |
| tomv-9 (2906–2932) | CCGTACCCTGCGCACTTTGCAAAatctacaacagtagaaattccctatagtgagtcgtattaatttc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alon, D.M.; Hak, H.; Bornstein, M.; Pines, G.; Spiegelman, Z. Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a. Plants 2021, 10, 1256. https://doi.org/10.3390/plants10061256
Alon DM, Hak H, Bornstein M, Pines G, Spiegelman Z. Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a. Plants. 2021; 10(6):1256. https://doi.org/10.3390/plants10061256
Chicago/Turabian StyleAlon, Dan Mark, Hagit Hak, Menachem Bornstein, Gur Pines, and Ziv Spiegelman. 2021. "Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a" Plants 10, no. 6: 1256. https://doi.org/10.3390/plants10061256
APA StyleAlon, D. M., Hak, H., Bornstein, M., Pines, G., & Spiegelman, Z. (2021). Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a. Plants, 10(6), 1256. https://doi.org/10.3390/plants10061256