Metabolomic Evaluation of Tissue-Specific Defense Responses in Tomato Plants Modulated by PGPR-Priming against Phytophthora capsici Infection
Abstract
:1. Introduction
2. Results
2.1. Symptom Development
2.2. Aromatic Amino Acid Quantification
2.3. Abscisic Acid (ABA) and Methyl Salicylic Acid (MeSA) Quantification
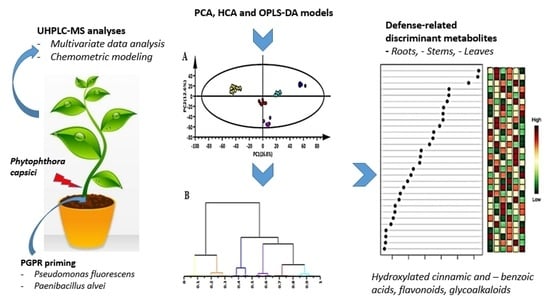
2.4. Metabolomic Profiling of PGPR-Primed Plants Responding to Phytophthora capsici
2.5. Time-Course of Comparative Metabolite Reprogramming in Primed-Unchallenged vs. Primed-Challenged Plants Inoculated with Phytophthora capsici
3. Discussion
4. Materials and Methods
4.1. Plant Material, PGPR Treatment and Infection with Phytophthora capsici
4.2. Experimental Design
4.3. Extraction and Quantification of Phytohormones and Aromatic Amino Acids by Multiple Reaction Monitoring Mass Spectrometry (MRM-MS)
Statistical Analysis of Amino Acid and Phytohormone Data
4.4. Metabolite Profiling: Sample Preparation, Analysis and Data Acquisition
4.5. Statistical Analyses of Multivariate Data (MVDA)
4.6. Metabolite Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2017, 23, 25–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant—Microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haichar, F.E.Z.; Santaella, C.; Heulin, T.; Achouak, W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014, 77, 69–80. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Dutta, S.; Podile, A.R. Plant growth promoting rhizobacteria (PGPR): The bugs to debug the root zone. Crit. Rev. Microbiol. 2010, 36, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 4, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, G.; Parihar, S.S.; Ahirwar, N.K.; Snehi, S.K.; Singh, V. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 7, 96–102. [Google Scholar]
- Ghosh, D.; Gupta, A.; Mohapatra, S. Dynamics of endogenous hormone regulation in plants by phytohormone secreting rhizobacteria under water-stress. Symbiosis 2019, 77, 265–278. [Google Scholar] [CrossRef]
- Sharma, S.; Chen, C.; Navathe, S.; Chand, R.; Pandey, S.P. A halotolerant growth promoting rhizobacteria triggers induced systemic resistance in plants and defends against fungal infection. Sci. Rep. 2019, 9, 4054. [Google Scholar] [CrossRef]
- Ortíz-Castro, R.; Valencia-Cantero, E.; López-Bucio, J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal. Behav. 2008, 3, 263–265. [Google Scholar] [CrossRef] [Green Version]
- Sanabria, N.M.; Huang, J.; Dubery, I.A. Self/non-self perception in plants in innate immunity and defense. Self Nonself Immune Recognit. Signal. 2009, 1, 40–54. [Google Scholar] [CrossRef] [Green Version]
- Shinya, T.; Motoyama, N.; Ikeda, A.; Wada, M.; Kamiya, K.; Hayafune, M.; Kaku, H.; Shibuya, N. Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012, 53, 1696–1706. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liu, J.; Ning, Y.; Ding, B.; Wang, X.; Wang, Z.; Wang, G.-L. Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant 2013, 6, 605–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.; Bheemaraddi, M.C.; Shivannavar, C.T. Biocontrol activity of siderophore producing Bacillus subtilis CTS-G24 against wilt and dry root rot causing fungi in chickpea. IOSR J. Agric. Vertin. Sci. 2014, 7, 63–68. [Google Scholar] [CrossRef]
- Ongena, M.; Duby, F.; Jourdan, E.; Beaudry, T.; Jadin, V.; Dommes, J.; Thonart, P. Bacillus subtilis M4 decreases plant susceptibility towards fungal pathogens by increasing host resistance associated with differential gene expression. Appl. Microbiol. Biotechnol. 2005, 67, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.L.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2015, 28, 984–995. [Google Scholar] [CrossRef] [Green Version]
- Schenk, S.T.; Hernández-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithöfer, A.; Becker, A.; Kogel, K.; et al. N-Acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schikora, A. Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Methods 2016, 90, 605–612. [Google Scholar] [CrossRef]
- Farag, M.A.; Zhang, H.; Ryu, C.-M. Dynamic chemical communication between plants and bacteria through airborne signals: Induced resistance by bacterial volatiles. J. Chem. Ecol. 2013, 39, 1007–1018. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.-S.; Yang, J.W.; Ryu, C.-M. ISR meets SAR outside: Additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front. Plant Sci. 2013, 4, 122. [Google Scholar] [CrossRef] [Green Version]
- Song, G.C.; Ryu, C.-M. Two Volatile organic compounds trigger plant self-defense against a bacterial pathogen and a sucking insect in cucumber under open field conditions. Int. J. Mol. Sci. 2013, 14, 9803–9819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the metabolic reconfiguration of the post-challenge primed state in Sorghum bicolor responding to Colletotrichum sublineolum infection. Metabolites 2019, 9, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempsey, D.A.; Klessig, D.F. SOS—Too many signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhage, A.; van Wees, S.C.M.; Pieterse, C.M.J. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Ocampo, L.M.; Vargas, A.M.; Naegele, R.P.; Francis, D.M.; Hausbeck, M.K. Resistance to crown and root rot caused by Phytophthora capsici in a tomato advanced backcross of Solanum habrochaites and Solanum lycopersicum. Plant Dis. 2015, 100, 829–835. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.K.; Kim, B.S.; Chang, S.W.; Hwang, B.K. Aggressiveness to pumpkin cultivars of isolates of Phytophthora capsici from pumpkin and pepper. Plant Dis. 2007, 85, 497–500. [Google Scholar] [CrossRef] [Green Version]
- Granke, L.L.; Quesada-Ocampo, L.; Lamour, K.; Hausbeck, M.K. Advances in research on Phytophthora capsici on vegetable crops in the United States. Plant Dis. 2012, 96, 1588–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laary, J.K. Dry-season farming and agrochemical misuse in upper east region of Ghana: Implication and way forward. J. Agric. Food Environ. Sci. 2012, 5, 1–10. [Google Scholar]
- Bhandari, G. An overview of agrochemicals and their effects on environment in Nepal. Appl. Ecol. Environ. Sci. 2014, 2, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Granke, L.L.; Windstam, S.T.; Hoch, H.C.; Smart, C.D.; Hausbeck, M.K. Dispersal and movement mechanisms of Phytophthora capsici sporangia. Phytopathology 2009, 99, 1258–1264. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Multi-platform metabolomic analyses of ergosterol-induced dynamic changes in Nicotiana tabacum cells. PLoS ONE 2014, 9, e87846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE 2016, 11, 0151350. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Concurrent metabolic profiling and quantification of aromatic amino acids and phytohormones in Solanum lycopersicum plants responding to Phytophthora capsici. Metabolites 2020, 10, 466. [Google Scholar] [CrossRef]
- Jupe, J.; Stam, R.; Howden, A.J.M.; Morris, J.A.; Zhang, R.; Hedley, P.E.; Huitema, E. Phytophthora capsici—Tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, R63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, P.G.; dos Santos, F.N.; Zanotta, S.; Eberlin, M.N.; Carazzone, C. Metabolomics of Solanum lycopersicum infected with Phytophthora infestans leads to early detection of late blight in asymptomatic plants. Molecules 2018, 23, 3330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed-Ab-Rahman, S.F.; Carvalhais, L.C.; Chua, E.; Xiao, Y.; Wass, T.J.; Schenk, P.M. Identification of soil bacterial isolates suppressing different Phytophthora spp. and promoting plant growth. Front. Plant Sci. 2018, 9, 1502. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. A chemometrics toolbox based on projections and latent variables. J. Chemom. 2014, 28, 332–346. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjo, L.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Sumner, L.W.; Samuel, T.; Noble, R.; Gmbh, S.D.; Barrett, D.; Beale, M.H.; Hardy, N. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2013, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G. The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. In The Arabidopsis Book; American Society of Plant Biologists (ASPB) Publication: Rockville, MD, USA, 2010; p. e0132. ISBN 1543-8120. [Google Scholar]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A three-ring circus: Metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Fagard, M.; Launay, A.; Clément, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulié, M.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Priming agents of plant defence stimulate the accumulation of mono- and di-acylated quinic acids in cultured tobacco cells. Physiol. Mol. Plant Pathol. 2014, 88, 61–66. [Google Scholar] [CrossRef]
- Mareya, C.R.; Tugizimana, F.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019, 9, 1840. [Google Scholar] [CrossRef] [Green Version]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic profiling of the host response of tomato (Solanum lycopersicum) following infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van Der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Shinkawa, R.; Morishita, A.; Amikura, K.; Machida, R.; Murakawa, H.; Kuchitsu, K.; Ishikawa, M. Abscisic acid induced freezing tolerance in chilling-sensitive suspension cultures and seedlings of rice. BMC Res. Notes 2013, 6, 351. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Medina, A.; Fernández, I.; Sánchez-Guzmán, M.J.; Jung, S.C.; Pascual, J.A.; Pozo, M.J. Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 2013, 4, 206. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; D’Auria, J.C.; Tholl, D.; Ross, J.R.; Gershenzon, J.; Noel, J.P.; Pichersky, E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003, 36, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718–721. [Google Scholar] [CrossRef]
- Manosalva, P.M.; Park, S.-W.; Forouhar, F.; Tong, L.; Fry, W.E.; Klessig, D.F. Methyl esterase 1 (StMES1) is required for systemic acquired resistance in potato. Mol. Plant Microbe Interact. 2010, 23, 1151–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K. Plant methyl salicylate induces defense responses in the rhizobacterium Bacillus subtilis. Environ. Microbiol. 2015, 17, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Chaturvedi, R.; Chowdhury, Z.; Venables, B.; Petros, R.A. Signaling by small metabolites in systemic acquired resistance. Plant J. 2014, 74, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamir, J.; Pastor, V.; Kaever, A.; Cerezo, M.; Flors, V. Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. Plant J. 2014, 78, 227–240. [Google Scholar] [CrossRef]
- Jang, S.; Ishihara, A.; Back, K. Production of coumaroylserotonin and feruloylserotonin in transgenic rice expressing pepper hydroxycinnamoyl-coenzyme A: Serotonin. Plant Physiol. 2004, 135, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Ly, D.; Kang, K.; Choi, J.-Y.; Ishihara, A.; Back, K.; Lee, S.-G. HPLC Analysis of serotonin, tryptamine, tyramine, and the hydroxycinnamic acid amides of serotonin and tyramine in food vegetables. J. Med. Food 2008, 11, 385–389. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Comparative metabolic phenotyping of tomato (Solanum lycopersicum) for the identification of metabolic signatures in cultivars differing in resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Labuschagne, N. Differential metabolic reprogramming in Paenibacillus alvei-primed Sorghum bicolor Seedlings in response to Fusarium pseudograminearum infection. Metabolites 2019, 9, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeiss, D.R.; Piater, L.A.; Dubery, I.A. Hydroxycinnamate amides: Intriguing conjugates of plant protective metabolites. Trends Plant Sci. 2021, 26, 184–195. [Google Scholar] [CrossRef]
- Cataldi, T.R.I.; Lelario, F.; Bufo, S.A. Analysis of tomato glycoalkaloids by liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 3103–3110. [Google Scholar] [CrossRef]
- Altesor, P.; García, Á.; Font, E.; Rodríguez-Haralambides, A.; Vilaró, F.; Oesterheld, M.; Soler, R.; González, A. Glycoalkaloids of wild and cultivated Solanum: Effects on specialist and generalist insect herbivores. J. Chem. Ecol. 2014, 40, 599–608. [Google Scholar] [CrossRef]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef]
- MetaboAnalyst—Statistical, Functional and Integrative Analysis of Metabolomics Data. Available online: www.metaboanalyst.ca (accessed on 21 August 2020).
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wishart, D.S. Using Metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 2016, 55, 14. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolomic Evaluation of Tissue-Specific Defense Responses in Tomato Plants Modulated by PGPR-Priming against Phytophthora capsici Infection. Plants 2021, 10, 1530. https://doi.org/10.3390/plants10081530
Mhlongo MI, Piater LA, Steenkamp PA, Labuschagne N, Dubery IA. Metabolomic Evaluation of Tissue-Specific Defense Responses in Tomato Plants Modulated by PGPR-Priming against Phytophthora capsici Infection. Plants. 2021; 10(8):1530. https://doi.org/10.3390/plants10081530
Chicago/Turabian StyleMhlongo, Msizi I., Lizelle A. Piater, Paul A. Steenkamp, Nico Labuschagne, and Ian A. Dubery. 2021. "Metabolomic Evaluation of Tissue-Specific Defense Responses in Tomato Plants Modulated by PGPR-Priming against Phytophthora capsici Infection" Plants 10, no. 8: 1530. https://doi.org/10.3390/plants10081530
APA StyleMhlongo, M. I., Piater, L. A., Steenkamp, P. A., Labuschagne, N., & Dubery, I. A. (2021). Metabolomic Evaluation of Tissue-Specific Defense Responses in Tomato Plants Modulated by PGPR-Priming against Phytophthora capsici Infection. Plants, 10(8), 1530. https://doi.org/10.3390/plants10081530