Functional Analysis of Jasmonates in Rice through Mutant Approaches
Abstract
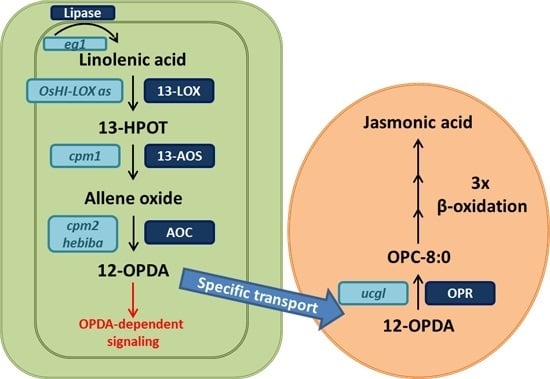
:1. Introduction
2. Rice Mutants and Transgenic Plants in the Jasmonate Pathway
3. Jasmonates and Their Link to Light
4. Jasmonates and Their Role in Rice Defense
5. Jasmonates and Their Relevance for the Response to Abiotic Stress
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takahashi, N.; Kitamura, H.; Kawarada, A.; Seta, Y.; Takai, M.; Tamura, S.; Sumiki, Y. Biochemical studies on “Bakanae” fungus. J. Agric. Chem. Soc. Japan 1955, 19, 267–281. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Marumo, S.; Ikekawa, N.; Morisaki, M.; Mori, K. Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 1981, 22, 323–325. [Google Scholar]
- Liu, Z.; Zhang, S.; Sun, N.; Liu, H.; Zhao, Y.; Liang, Y.; Zhang, L.; Han, Y. Functional diversity of jasmonates in rice. Rice 2015. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.K.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef] [PubMed]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in arabidopsis leaves. J. Biol. Chem. 2013, 288, 31701–31714. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Falkenstein, E.; Groth, B.; Mithöfer, A.; Weiler, E. Methyljasmonate and α-linolenic acid are potent inducers of tendril coiling. Planta 1991, 185, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Svyatyna, K.; Riemann, M. Light-dependent regulation of the jasmonate pathway. Protoplasma 2012, 249, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Iino, M. Phytochrome-mediated transcriptional up-regulation of ALLENE OXIDE SYNTHASE in rice seedlings. Plant Cell Physiol. 2004, 45, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Oshima, T.; Kaneda, H.; Takashio, M. Identification and functional analyses of two cDNAs that encode fatty acid 9-/13-hydroperoxide lyase (CYP74C) in rice. Biosci. Biotechnol. Biochem. 2005, 69, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Muller, A.; Korte, A.; Furuya, M.; Weiler, E.W.; Nick, P. Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 2003, 133, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Nordström, K.J.V.; Albani, M.C.; James, G.V.; Gutjahr, C.; Hartwig, B.; Turck, F.; Paszkowski, U.; Coupland, G.; Schneeberger, K. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 2013, 31, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M.G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S.Y.; Nadal, M.; et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.K.; Neumann, R.; Haga, K.; Yatoh, O.; Iino, M. Photomorphogenesis of rice seedlings: A mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol. 2003, 44, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Kiyota, S.; Jikumaru, Y.; Kamiya, Y.; Takano, M.; Iino, M. Functional analysis of a rice allene oxide synthase gene (OsAOS1) that functions for jasmonate biosynthesis. Plant Cell Physiol. Suppl. 2008. [Google Scholar] [CrossRef]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Qi, J.; Ren, N.; Cheng, J.; Erb, M.; Mao, B.; Lou, Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009, 60, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Robert, C.A.M.; Riemann, M.; Cosme, M.; Mène-Saffrané, L.; Massana, J.; Stout, M.J.; Lou, Y.; Gershenzon, J.; Erb, M. Induced jasmonate signaling leads to contrasting effects on root damage and herbivore performance. Plant Physiol. 2015, 167, 1100–1116. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Shi, C.; Zeng, D.; Jin, X.; Wu, J. Morphogenesis and molecular basis on the unclosed glumes, a novel mutation related to the floral organ of rice. Plant Mol. Biol. Report. 2015, 33, 480–489. [Google Scholar] [CrossRef]
- Tani, T.; Sobajima, H.; Okada, K.; Chujo, T.; Arimura, S.I.; Tsutsumi, N.; Nishimura, M.; Seto, H.; Nojiri, H.; Yamane, H. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 2008, 227, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Riemann, M.; Takano, M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant Cell Environ. 2008, 31, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chen, Y.; Charnikhova, T.; Mulder, P.P.J.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.B.; Dreni, L.; et al. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Abe, H.; Arimura, G.I. Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol. 2015, 56, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Seo, J.S.; Cho, J.H.; Jung, H.; Kim, J.K.; Lee, J.S.; Rhee, S.; do Choi, Y. Oryza sativa COI Homologues Restore Jasmonate Signal Transduction in Arabidopsis coi1-1 Mutants. PLoS ONE 2013, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. PNAS Plus: Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Sakuraba, Y.; Lee, T.; Kim, K.W.; An, G.; Lee, H.Y.; Paek, N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef] [PubMed]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H.; Yamakawa, H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Gutjahr, C.; Korte, A.; Danger, B.; Muramatsu, T.; Bayer, U.; Waller, F.; Furuya, M.; Nick, P. GER1, a GDSL motif-encoding gene from rice is a novel early light- and jasmonate-induced gene. Plant Biol. 2007, 9, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.L.; Okamoto, H.; Wang, M.; Ang, L.H.; Matsui, M.; Goodman, H.; Deng, X.W. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 2000, 14, 1958–1970. [Google Scholar] [PubMed]
- Chen, I.C.; Huang, I.C.; Liu, M.J.; Wang, Z.G.; Chung, S.S.; Hsieh, H.L. Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 2007, 143, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Okamoto, H.; Patrick, E.; Harris, S.R.; Wasternack, C.; Brearley, C.; Turner, J.G. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 2010, 22, 1143–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Li, C.B.; Zheng, W.; Wu, X.; Zhao, J.; Zhou, G.; Jiang, H.; Sun, J.; Lou, Y.; Li, C. Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol. 2007, 48, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sonobe, K.; Ogawa, N.; Masuda, S.; Nagatani, A.; Kobayashi, Y.; Ohta, H. Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J. Plant Res. 2013, 126, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. The interplay between light and jasmonate signalling during defence and development. J. Exp. Bot. 2011, 62, 4087–4100. [Google Scholar] [CrossRef] [PubMed]
- Sineshchekov, V.A.; Loskovich, A.V.; Riemann, M.; Nick, P. The jasmonate-free rice mutant hebiba is affected in the response of phyA’/phyA” pools and protochlorophyllide biosynthesis to far-red light. Photochem. Photobiol. Sci. 2004, 3, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Bouyer, D.; Hisada, A.; Müller, A.; Yatou, O.; Weiler, E.W.; Takano, M.; Furuya, M.; Nick, P. Phytochrome A requires jasmonate for photodestruction. Planta 2009, 229, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Brendel, R.; Svyatyna, K.; Jikumaru, Y.; Reichelt, M.; Mithöfer, A.; Takano, M.; Kamiya, Y.; Nick, P.; Riemann, M. Effects of light and wounding on jasmonates in rice phyAphyC mutants. Plants 2014, 3, 143–159. [Google Scholar] [CrossRef]
- Svyatyna, K.; Jikumaru, Y.; Brendel, R.; Reichelt, M.; Mithöfer, A.; Takano, M.; Kamiya, Y.; Nick, P.; Riemann, M. Light induces jasmonate-isoleucine conjugation via OsJAR1-dependent and -independent pathways in rice. Plant Cell Environ. 2014, 37, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Wakuta, S.; Suzuki, E.; Saburi, W.; Matsuura, H.; Nabeta, K.; Imai, R.; Matsui, H. OsJAR1 and OsJAR2 are jasmonyl-l-isoleucine synthases involved in wound- and pathogen-induced jasmonic acid signalling. Biochem. Biophys. Res. Commun. 2011, 409, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.L.; Kang, J.H.; Howe, G.A. Jasmonate-triggered plant immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Gheysen, G.; Höfte, M. Hormone defense networking in rice: Tales from a different world. Trends Plant Sci. 2013, 18, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Höfte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Luo, S.M.; Xie, J.F.; Li, Y.F.; Xu, T.; Liu, Y.; Song, Y.Y.; Zhu-Salzman, K.; Zeng, R. Sen Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense. PLoS ONE 2012, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Dubugnon, L. Detritivorous crustaceans become herbivores on jasmonate-deficient plants. Proc. Natl. Acad. Sci. USA 2009, 106, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Ribot, C.; Hirsch, J.; Balzergue, S.; Tharreau, D.; Nottéghem, J.L.; Lebrun, M.H.; Morel, J.B. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 2008, 165, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.J.; Valent, B. Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 1996, 50, 491–512. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.; Buchala, A.; Metraux, J.P. Gene-expression patterns and levels of jasmonic acid in rice treated with the resistance inducer 2,6-dichloroisonicotinic acid. Plant Physiol. 1997, 115, 61–70. [Google Scholar] [PubMed]
- Schweizer, P.; Buchala, A.; Silverman, P.; Seskar, M.; Raskin, I.; Metraux, J.P. Jasmonate-inducible genes are activated in rice by pathogen attack without a concomitant increase in endogenous jasmonic acid levels. Plant Physiol. 1997, 114, 79–88. [Google Scholar] [PubMed]
- Shimizu, T.; Miyamoto, K.; Miyamoto, K.; Minami, E.; Nishizawa, Y.; Iino, M.; Nojiri, H.; Yamane, H.; Okada, K. OsJAR1 contributes mainly to biosynthesis of the stress-induced jasmonoyl-isoleucine involved in defense responses in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Huffaker, A.; Sims, J.W.; Christensen, S.A.; Lu, X.; Okada, K.; Peters, R.J. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014, 79, 659–678. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Lin, F.; Hasegawa, M.; Okada, K.; Nojiri, H.; Yamane, H. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 2012, 287, 19315–19325. [Google Scholar] [CrossRef] [PubMed]
- Patkar, R.N.; Benke, P.I.; Qu, Z.; Constance Chen, Y.Y.; Yang, F.; Swarup, S.; Naqvi, N.I. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat. Chem. Biol. 2015, 11, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Li, J.; Zhou, G.; Wang, Q.; Bian, W.; Erb, M.; Lou, Y. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. Elife 2015, 4, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, J.; Peng, X.; Tang, X.; Zhou, P.; Xiang, J.; Deng, X. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 2015, 89, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; Takahashi, A.; Nishizawa, Y.; Minami, E.; Nojiri, H.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 2014, 9, e98737. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Shimogawa, T.; Shimizu, T.; Otake, Y.; Yokotani, N.; Nishizawa, Y.; Shibuya, N.; Nojiri, H.; Yamane, H.; et al. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 2013, 82, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring jasmonates in the hormonal network of drought and salinity responses. Front. Plant Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Pedranzani, H.; Racagni, G.; Alemano, S. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003, 41, 149–158. [Google Scholar] [CrossRef]
- Moons, A.; Prinsen, E.; Bauw, G.; van Montagu, M. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 1997, 9, 2243–2259. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.J.; Seo, Y.J.; Lee, J.D.; Ishii, R.; Kim, K.U.; Shin, D.H.; Park, S.K.; Jang, S.W.; Lee, I.J. Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Sci. 2005, 191, 273–282. [Google Scholar] [CrossRef]
- Ismail, A.; Seo, M.; Takebayashi, Y.; Kamiya, Y.; Eiche, E.; Nick, P. Salt adaptation requires efficient fine-tuning of jasmonate signalling. Protoplasma 2014, 251, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Takeda, S.; Nick, P. Life and death under salt stress: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 2015, 66, 3339–3352. [Google Scholar] [CrossRef] [PubMed]
- Kurotani, K.I.; Hayashi, K.; Hatanaka, S.; Toda, Y.; Ogawa, D.; Ichikawa, H.; Ishimaru, Y.; Tashita, R.; Suzuki, T.; Ueda, M.; et al. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015, 56, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, N.; Matsubara, T.; Sato, M.; Takahashi, K.; Wakuta, S.; Kawaide, H.; Matsui, H.; Nabeta, K.; Matsuura, H. Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011, 52, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, M.A.; Stingl, N.E.; Lautenschlaeger, J.K.; Krischke, M.; Mueller, M.J.; Berger, S. Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 2010, 152, 1940–1950. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, M.A.; Hussels, W.; Berger, S. Jasmonates during senescence: Signals or products of metabolism? Plant Signal. Behav. 2010, 5, 1493–1496. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [PubMed]
- Savchenko, T.; Kolla, V.; Wang, C.Q.; Nasafi, Z.; Hicks, D.; Phadungchob, B.; Chehab, W.; Brandizzi, F.; Froehlich, J.; Dehesh, K. Functional convergence of oxylipin and ABA pathways controls stomatal closure in response to drought. Plant Physiol. 2014, 164, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Choi, Y.D.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, M.; Brady, S.M. Gene regulatory networks in the Arabidopsis root. Curr. Opin. Plant Biol. 2013, 16, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arbaizar, A.; Regalado, J.J.; Lorenzo, O. Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the arabidopsis coronatine insensitive 1-16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol. 2012, 53, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Bray, E.A. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 2006, 57, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.Y.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Seah, S.W.; Xu, J. The root of ABA action in environmental stress response. Plant Cell Rep. 2013, 32, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Gyohda, A.; Tominaga, M.; Kawakatsu, M.; Hatakeyama, A.; Ishii, N.; Shimaya, K.; Nishimura, T.; Riemann, M.; Nick, P.; et al. RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol. 2011, 52, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Arbona, V.; Argamasilla, R.; Gómez-Cadenas, A. Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. J. Plant Physiol. 2010, 167, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- De Ollas, C.; Hernando, B.; Arbona, V.; Gómez-Cadenas, A. Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiol. Plant. 2013, 147, 296–306. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | RAP ID | MSU ID | Function | Reference |
|---|---|---|---|---|
| OsJAZ1, OsTIFY3 | Os04g0653000 | LOC_Os04g55920 | Floral development | [29] |
| OsJAZ8, OsTIFY10c | Os09g0439200 | LOC_Os09g26780 | Plant defense | [40] |
| OsJAZ9, OsTIFY11a | Os03g0180800 | LOC_Os03g08310 | Salinity stress | [38,39] |
| OsJAZ10, OsTIFY11b | Os03g0181100 | LOC_Os03g08330 | Stem growth and grain size | [41] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhakarey, R.; Kodackattumannil Peethambaran, P.; Riemann, M. Functional Analysis of Jasmonates in Rice through Mutant Approaches. Plants 2016, 5, 15. https://doi.org/10.3390/plants5010015
Dhakarey R, Kodackattumannil Peethambaran P, Riemann M. Functional Analysis of Jasmonates in Rice through Mutant Approaches. Plants. 2016; 5(1):15. https://doi.org/10.3390/plants5010015
Chicago/Turabian StyleDhakarey, Rohit, Preshobha Kodackattumannil Peethambaran, and Michael Riemann. 2016. "Functional Analysis of Jasmonates in Rice through Mutant Approaches" Plants 5, no. 1: 15. https://doi.org/10.3390/plants5010015
APA StyleDhakarey, R., Kodackattumannil Peethambaran, P., & Riemann, M. (2016). Functional Analysis of Jasmonates in Rice through Mutant Approaches. Plants, 5(1), 15. https://doi.org/10.3390/plants5010015