Genetic Variation in Damaged Populations of Pistacia atlantica Desf.
Abstract
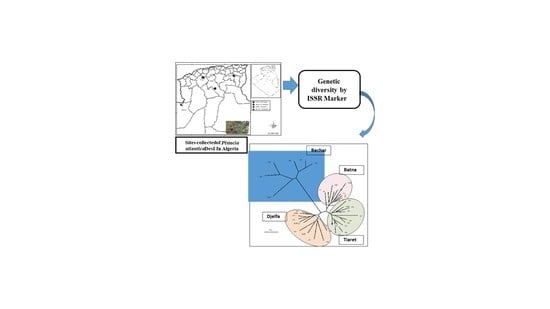
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction
4.3. ISSR-PCR Amplification
4.4. Data Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zohary, M. A monographical study of the genus Pistacia. Palestine J. Bot. 1952, 5, 187–228. [Google Scholar]
- Labdelli, A.; Zemour, K.; Simon, V.; Cerny, M.; Adda, A.; Merah, O. Pistacia atlantica Desf., a Source of Healthy Vegetable Oil. Appl. Sci. 2019, 9, 2552. [Google Scholar] [CrossRef] [Green Version]
- Labdelli, A.; Rebiai, A.; Tahirine, M.; Adda, A.; Merah, O. Nutritional Content and Antioxidant Capacity of the Seed and the Epicarp in Different Ecotypes of Pistacia atlantica Desf. subsp. atlantica. Plants 2020, 9, 1065. [Google Scholar] [CrossRef] [PubMed]
- Ben Ahmed, Z.; Yousfi, M.; Viaene, J.; Dejaegher, B.; Demeyer, K.; Heyden, Y.V. Four Pistacia atlantica subspecies (atlantica, cabulica, kurdica and mutica): A review of their botany, ethnobotany, phytochemistry and pharmacology. J. Ethnopharmacol. 2020, 265, 113329. [Google Scholar] [CrossRef] [PubMed]
- Belhadj, S.; Derridj, A.; Gauquelin, T. Pistachio use in Algeria. In: Avanzato D, Vassallo I. (eds) Following pistachio footprints (Pistacia vera L.). Cultivation and culture, folklore and history, traditions and uses. Script. Hortic. 2008, 7, 15–21. [Google Scholar]
- El Zerey-Belaskri, A. Taxonomic and botanical retrospective review of Pistacia atlantica Desf. (Anacardiaceae). Arab. J. Med. Aromat. Plants 2019, 5, 47–77. [Google Scholar] [CrossRef]
- Said, S.A.; Fernandez, C.; Greff, S.; Derridj, A.; Gauquelin, T.; Mevy, J.-P. Inter-population variability of leaf morpho-anatomical and terpenoid patterns of Pistacia atlantica Desf. ssp. atlantica growing along an aridity gradient in Algeria. Flora Morphol. Distrib. Funct. Ecol. Plants 2011, 206, 397–405. [Google Scholar] [CrossRef]
- Amara, M.; Bouazza, M.; Al Saghir, B.A.M. Anatomical and Adaptation Features of Pistacia atlantica Desf. to Adverse Climate Conditions in Algeria. Am. J. Plant Sci. 2017, 8, 137–153. [Google Scholar] [CrossRef] [Green Version]
- Zerey-Belaskri, E.; Benhassaini, H. Morphological leaf variability in natural populations of Pistacia atlantica Desf. subsp. atlantica along climatic gradient: New features to update Pistacia atlantica subsp. atlantica key. Int. J. Biometeorol. 2015, 60, 577–589. [Google Scholar] [CrossRef]
- Al-Saghir, M.G.; Porter, D.M. Taxonomic Revision of the Genus Pistacia L. (Anacardiaceae). Am. J. Plant Sci. 2012, 3, 12–32. [Google Scholar] [CrossRef] [Green Version]
- Shahghobadi, H.; Shabanian, N.; Rahmani, M.-S.; Khadivi-Khub, A. Genetic characterization of Pistacia atlantica subsp. kurdica from northern Zagros forests in Iran. Trees 2018, 33, 481–490. [Google Scholar] [CrossRef]
- Xie, L.; Yang, Z.-Y.; Wen, J.; Li, D.-Z.; Yi, T.-S. Biogeographic history of Pistacia (Anacardiaceae), emphasizing the evolution of the Madrean-Tethyan and the eastern Asian-Tethyan disjunctions. Mol. Phylogenet. Evol. 2014, 77, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Golan-Goldhirsh, A.; Barazani, O.; Wang, Z.S.; Khadka, D.; Saunders, J.A.; Kostiukovsky, V.; Rowland, L.J. Genetic relationships among Mediterranean Pistacia species evaluated by RAPD and AFLP markers. Plant Syst. Evol. 2004, 246, 9–18. [Google Scholar] [CrossRef]
- Arefi, H.; Abdi, A.; Saydian, S.; Nasirzadeh, A.; Nadushan, H.; Rad, M.; Golbabaii, H.; Azdoo, Z.; Ziedabadi, D. Genetics and breeding of pistacia atlantica in Iran. Acta Hortic. 2006, 726, 77–84. [Google Scholar] [CrossRef]
- Karimi, H.R.; Kafkas, S. Genetic relationships among Pistacia species studied by SAMPL markers. Plant Syst. Evol. 2011, 297, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Kafkas, S.; Perl-Treves, R. Morphological and molecular phylogeny of Pistacia species in Turkey. Theor. Appl. Genet. 2001, 102, 908–915. [Google Scholar] [CrossRef]
- Katsiotis, A.; Hagidimitriou, M.; Drossou, A.; Pontikis, C.; Loukas, M. Genetic relationships among species and cultivars of Pistacia using RAPDs and AFLPs. Euphytica 2003, 132, 279–286. [Google Scholar] [CrossRef]
- Mirzaei, S.; Bahar, M.; Sharifnabi, B. A phylogenetic study of iranian wild pistachio species and some cultivars using rapd markers. Acta Hortic. 2006, 726, 39–44. [Google Scholar] [CrossRef]
- El Zerey-Belaskria, A.; Ribeirob, T.; Alcarazc, M.L.; El Zereyd, W.; Castroe, S.; Loureiroe, J.; Benhassainia, H.; Hormaza, J.I. Molecular characterization of Pistacia atlantica Desf. subsp. atlantica (Anacardiaceae) in Algeria: Genome size determination, chromosome count and genetic diversity analysis using SSR markers. Sci. Hortic. 2018, 227, 278–287. [Google Scholar] [CrossRef]
- Basha, A.I.; Padulosi, S.; Chabane, K.; Hadj-Hassan, A.; Dulloo, E.; Pagnotta, M.A.; Porceddu, E. Genetic diversity of Syrian pistachio (Pistacia vera L.) varieties evaluated by AFLP markers. Genet. Resour. Crop Evol. 2007, 54, 1807–1816. [Google Scholar] [CrossRef]
- Pazouki, L.; Mardi, M.; Shanjani, P.S.; Hagidimitriou, M.; Pirseyedi, S.M.; Naghavi, M.R.; Avanzato, D.; Vendramin, E.; Kafkas, S.; Ghareyazie, B.; et al. Genetic diversity and relationships among Pistacia species and cultivars. Conserv. Genet. 2010, 11, 311–318. [Google Scholar] [CrossRef]
- Arabnezhad, H.; Bahar, M.; Pour, A.T. Evaluation of genetic relationships among Iranian pistachios using microsatellite markers developed from Pistacia khinjuk Stocks. Sci. Hortic. 2011, 128, 249–254. [Google Scholar] [CrossRef]
- Motalebipour, E.Z.; Kafkas, S.; Khodaeiaminjan, M.; Çoban, N.; Gözel, H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: Development of novel SSR markers and genetic diversity in Pistacia species. BMC Genom. 2016, 17, 998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khadivi-Khub, A.; Esmaeili, A.; Mardani, N. Genetic diversity of cultivated pistachio as revealed by microsatellite molecular markers. Biotechnol. Biotechnol. Equip. 2018, 32, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Kafkas, S.; Ozkan, H.; Ak, B.E.; Acar, I.; Atli, H.S.; Koyuncu, S. Detecting DNA Polymorphism and Genetic Diversity in a Wide Pistachio Germplasm: Comparison of AFLP, ISSR, and RAPD Markers. J. Am. Soc. Hortic. Sci. 2006, 131, 522–529. [Google Scholar] [CrossRef]
- Pourian, M.A.; Bakhshi, D.; Aalami, A.; Hokmabadi, H. Assessment of Genetic Relationships among Cultivated and Wild Pistachios (Pistacia vera L.) using Molecular Markers. J. Hortic. Res. 2019, 27, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Chen, F.; Yeh, K.-W.; Chen, J. ISSR Analysis of Genetic Diversity and Structure of Plum Varieties Cultivated in Southern China. Biology 2018, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Fares, K.; Guasmi, F.; Touil, L.; Triki, T.; Ferchichi, A. Genetic Diversity of Pistachio Tree using Inter-Simple Sequence Repeat Markers ISSR Supported by Morphological and Chemical Markers. Biotechnology 2009, 8, 24–34. [Google Scholar] [CrossRef]
- Tagizad, A.; Ahmadi, J.; Haddad, R.; Zarrabi, M. A comparative analysis of ISSR and RAPD markers for studying genetic diversity in Iranian pistachio cultivars. IJGPB 2010, 1, 6–16. [Google Scholar]
- Turkeli, Y.; Kafkas, S. First genetic linkage map in pistachio constructed using an interspecific cross between Pistacia vera L. and monoecious Pistacia atlantica Desf. Sci. Hortic. 2013, 151, 30–37. [Google Scholar] [CrossRef]
- Gupta, M.; Chyi, Y.S.I.; Romero-Severson, J.; Owen, J.L. Amplification of DNA markers from evolutionary diverse genomes using single primers of simple sequence repeats. Theor. Appl. Genet. 1994, 89, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358. [Google Scholar] [CrossRef] [PubMed]
- Arcade, A.; Anselin, F.; Rampant, P.F.; Lesage, M.C.; Pâques, L.E.; Prat, D. Application of AFLP, RAPD and ISSR markers to genetic mapping of European and Japanese larch. Theor. Appl. Genet. 2000, 100, 299–307. [Google Scholar] [CrossRef]
- Goulão, L.; Valdiviesso, T.; Santana, C.; Oliveira, C.M. Comparison between phenetic characterization using RAPD and ISSR markers and phenotypic data of cultivated chestnut (Castanea sativa Mill.). Genet. Resour. Crop Evol. 2001, 48, 329–338. [Google Scholar] [CrossRef]
- Quézel, P.; Médail, F.; Loisel, R.; Barbero, M. Biodiversity and conservation of forest species in the Mediterranean basin. Unasylva 1999, 50, 21–28. [Google Scholar]
- Kafkas, S. Phylogenetic analysis of the genus Pistacia by AFLP markers. Plant Syst. Evol. 2006, 262, 113–124. [Google Scholar] [CrossRef]
- Al-Sousli, M.; Faory, H.; Nakar, M.; Zaid, S.; Al-Safadi, B.; Al-Saghir, M. Genetic relationships among some Pistacia species (Anacardiaceae) in Syria. Middle East J. Sci. Res. 2014, 21, 1487–1496. [Google Scholar] [CrossRef]
- Schlüter, P.M.; Harris, S.A. Analysis of multilocus fingerprinting data sets containing missing data. Mol. Ecol. Notes 2006, 6, 569–572. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 1993, 47, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]




| Primer | Batna | Djelfa | Tiaret | Bechar | |
|---|---|---|---|---|---|
| ISSR3 | Total number of bands | 19 | 14 | 19 | 13 |
| Number of polymorphic bands | 13 | 7 | 14 | 9 | |
| % polymorphic bands | 68.42 | 50 | 73.68 | 69.23 | |
| ISSR4 | Total number of bands | 14 | 14 | 13 | 6 |
| Number of polymorphic bands | 8 | 7 | 10 | 4 | |
| % polymorphic bands | 57.14 | 50 | 76.92 | 66.67 |
| Populations | Na | %P | Nd | Ne | He | SI |
|---|---|---|---|---|---|---|
| Batna | 83 | 63.64% | 1.636 ± 0.085 | 1.504 ± 0.076 | 0.272 ± 0.039 | 5.26 ± 0.29 |
| Djelfa | 83 | 50% | 1.500 ± 0.096 | 1.440 ± 0.086 | 0.233 ± 0.045 | 5.34 ± 0.36 |
| Tiaret | 83 | 75% | 1.750 ± 0.078 | 1.640 ± 0.069 | 0.344 ± 0.036 | 5.32 ± 0.17 |
| Bechar | 33 | 68.42% | 1.684 ± 0.110 | 1.565 ± 0.071 | 0.305 ± 0.051 | 3.6 ± 0.19 |
| Assigned Code | Location | Number of Samples | Altitude (m) | Latitude (N) | Longitude |
|---|---|---|---|---|---|
| B | Batna Semi-arid | 10 | 1027 | 35°37′10″ | 6°22′13″ E |
| T | Tiaret Semi-arid | 12 | 808 | 35°22′33″ | 02°09′5″ W |
| D | Djelfa Arid | 11 | 630 | 34°02′11″ | 03°40′22″ E |
| A | Bechar Hyperarid | 06 | 979 | 32°04′6″ | 02°18′5″ W |
| Primer Name | Sequence 5′–3′ | Annealing Temperature (°C) |
|---|---|---|
| ISSR 1 | (CG)9W | From 50 to 60 |
| ISSR 2 | (GC)9W | From 50 to 60 |
| ISSR 3 | (AG)9B | 54 |
| ISSR 4 | (GA)9Y | 54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labdelli, A.; De La Herrán, R.; Arafeh, R.; Resentini, F.; Trainotti, L.; Halis, Y.; Adda, A.; Merah, O. Genetic Variation in Damaged Populations of Pistacia atlantica Desf. Plants 2020, 9, 1541. https://doi.org/10.3390/plants9111541
Labdelli A, De La Herrán R, Arafeh R, Resentini F, Trainotti L, Halis Y, Adda A, Merah O. Genetic Variation in Damaged Populations of Pistacia atlantica Desf. Plants. 2020; 9(11):1541. https://doi.org/10.3390/plants9111541
Chicago/Turabian StyleLabdelli, Amina, Roberto De La Herrán, Rami Arafeh, Francesca Resentini, Livio Trainotti, Youcef Halis, Ahmed Adda, and Othmane Merah. 2020. "Genetic Variation in Damaged Populations of Pistacia atlantica Desf." Plants 9, no. 11: 1541. https://doi.org/10.3390/plants9111541
APA StyleLabdelli, A., De La Herrán, R., Arafeh, R., Resentini, F., Trainotti, L., Halis, Y., Adda, A., & Merah, O. (2020). Genetic Variation in Damaged Populations of Pistacia atlantica Desf. Plants, 9(11), 1541. https://doi.org/10.3390/plants9111541