Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mucus Collection and Separation of Different Fractions
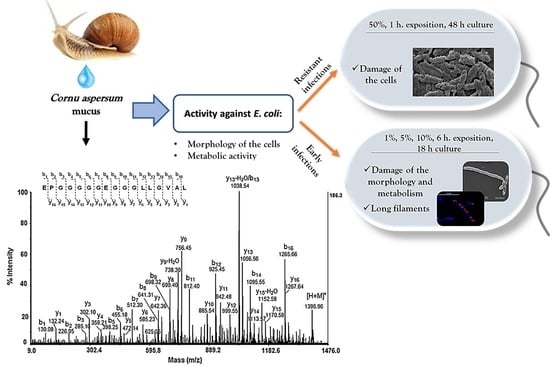
2.2. Analysis of Peptide Fractions by Mass Spectrometric Analysis
2.3. Antibacterial Activity Testing
2.4. Nutrient Media and Culture Conditions
2.5. Studies of Antibacterial Activities Using Different Methods
2.6. Electron Microscopic Assays
2.7. CTC/DAPI Staining and Digital Image Analysis
3. Results
3.1. Analysis and Physicochemical Characteristics of the Fraction
3.2. The Antibacterial Activity—24 h Impact on Cells of E. coli in a Late Logarithmic Phase in a Solid Nutrient Media with Deep and Surface Inoculations of the Bacterial Material
3.3. Antibacterial Activity—First Variant
3.4. Antibacterial Activity—Second Variant
3.5. Antibacterial Activity—Third Variant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaslof, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef]
- Joo, H.S.; Fu, C.I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos Trans. R. Soc. B Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Mandel, S.; Michaeli, J.; Nur, N.; Erbetti, I.; Zazoun, J.; Ferrari, L.; Felici, A.; Cohen-Kutner, M.; Bachnoff, N. OMN6 a novel bioengineered peptide for the treatment of multidrug resistant Gram negative bacteria. Sci. Rep. 2021, 11, 6603. [Google Scholar] [CrossRef] [PubMed]
- Witherell, K.S.; Price, J.; Bandaranayake, A.D.; Olson, J.; Call, D.R. In vitro activity of antimicrobial peptide CDP-B11 alone and in combination with colistin against colistin-resistant and multidrug-resistant Escherichia coli. Sci. Rep. 2021, 11, 2151. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Berisio, R.; Grieco, P.; Morelli, G.; Galdiero, M. Antimicrobial peptides as an opportunity against bacterial diseases. Curr. Med. Chem. 2015, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Ebbensgaard, A.; Mordhorst, H.; Overgaard, M.T.; Nielsen, C.G.; Aarestrup, F.M.; Hansen, E.B. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS ONE 2015, 10, e0144611. [Google Scholar] [CrossRef] [Green Version]
- Lima, B.; Ricci, M.; Garro, A.; Juhász, T.; Szigyártó, I.C.; Papp, Z.I.; Feresin, G.; de la Torre, J.G.; Cascales, J.L.; Fülöp, L.; et al. New short cationic antibacterial peptides. Synthesis, biological activity and mechanism of action. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183665. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Securing New Drugs for Future Generations: The Pipeline of Antibiotics. (The Review on Antimicrobial Resistance, 2015). Available online: https://amr-review.org/sites/default/files/SECURING%20NEW%20DRUGS%20FOR%20FUTURE%20GENERATIONS%20FINAL%20WEB_0.pdf (accessed on 10 January 2022).
- World Health Organization. Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use, 5th ed.; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Totsika, M. Benefits and challenges of antivirulence antimicrobials at the dawn of the post-antibiotic era. Curr. Med. Chem. 2016, 6, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, J.; Coates, C.J.; Zhu, H.; Zhu, P.; Zujian, W.; Xie, L. Identification of candidate antimicrobial peptides derived from abalone hemocyanin. Dev. Comp. Immunol. 2015, 49, 96–102. [Google Scholar] [CrossRef]
- Li, H.; Parisi, M.G.; Parrinello, N.; Cammarata, M.; Roch, P. Molluscan antimicrobial peptides, a review from activity-based evidences to computer-assisted sequences. Invertebr. Surviv. J. 2011, 8, 85–97. [Google Scholar]
- Pitt, S.; Graham, M.A.; Dedi, C.G.; Taylor-Harris, P.M.; Gunn, A. Antimicrobial properties of mucus from the brown garden snail Helix aspersa. Br. J. Biomed. Sci. 2015, 72, 174–181. [Google Scholar] [CrossRef] [Green Version]
- González García, M.; Rodríguez, A.; Alba, A.; Vázquez, A.A.; Morales Vicente, F.E.; Pérez-Erviti, J.; Spellerberg, B.; Stenger, S.; Grieshober, M.; Conzelmann, C.; et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927). Biomolecules 2020, 10, 1473. [Google Scholar] [CrossRef]
- Wang, L.; Qiu, L.; Zhou, Z.; Song, L. Research progress on the mollusc immunity in China. Dev. Comp. Immunol. 2013, 39, 2–10. [Google Scholar] [CrossRef]
- Dolashka, P.; Moshtanska, V.; Borisova, V.; Dolashki, A.; Stevanovic, S.; Dimanov, T.; Voelter, W. Antimicrobial proline-rich peptides from the hemolymph of marine snail Rapana venosa. Peptides 2011, 32, 1477. [Google Scholar] [CrossRef]
- Dolashka, P.; Dolashki, A.; Velkova, L.; Stevanovic, S.; Molin, L.; Traldi, P.; Velikova, R.; Voelter, W. Bioactive compounds isolated from garden Snails. J. BioSci. Biotechnol. 2015, 147–155. [Google Scholar]
- Leoni, G.; De Poli, A.; Mardirossian, M.; Gambato, S.; Florian, F.; Venier, P.; Wilson, D.N.; Tossi, A.; Pallavicini, A.; Gerdol, M. Myticalins: A novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.). Mar. Drugs. 2017, 15, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassone, M.; Otvos, L., Jr. Synergy among antibacterial peptides and between peptides and small-molecule antibiotics. Expert Rev. Anti-Infect. Ther. 2010, 8, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolashka, P.; Dolashki, A.; Voelter, W.; Beeumen, J.; Stevanovic, S. Antimicrobial activity of peptides from the hemolymph of Helix lucorum snails. Int. J. Curr. Microbiol. App. Sci. 2015, 4, 1061–1071. [Google Scholar]
- Zhong, J.; Wang, W.; Yang, X.; Yan, X.; Liu, R. A novel cysteine-rich antimicrobial peptide from the mucus of the snail of Achatina fulica. Peptides 2013, 39, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Suárez, L.; Pereira, A.; Hidalgo, W.; Uribe, N. Antibacterial, Antibiofilm and Anti-Virulence Activity of Biactive Fractions from Mucus Secretion of Giant African Snail Achatina fulica against Staphylococcus aureus Strains. Antibiotics 2021, 10, 1548. [Google Scholar] [CrossRef]
- Smith, A.M.; Quick, T.J.; St Peter, R.L. Differences in the Composition of Adhesive and Non-Adhesive Mucus from the Limpet Lottia limatula. Biol. Bull. 1999, 196, 34–44. [Google Scholar] [CrossRef]
- Pitt, S.J.; Hawthorne, J.A.; Garcia-Maya, M.; Alexandrovich, A.; Symonds, R.C.; Gunn, A. Identification and characterisation of anti-Pseudomonas aeruginosa proteins in mucus of the brown garden snail, Cornu aspersum. Br. J. Biomed. Sci. 2019, 76, 129–136. [Google Scholar] [CrossRef]
- Kubota, Y.; Watanabe, Y.; Otsuka, H.; Tamiya, T.; Tsuchiya, T.; Matsumoto, J.J. Purification and characterization of an antibacterial factor from snail mucus. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 1985, 82, 345–348. [Google Scholar] [CrossRef]
- Ulagesan, S.; Kim, H.J. Antibacterial and Antifungal Activities of Proteins Extracted from Seven Different Snails. Appl. Sci. 2018, 8, 1362. [Google Scholar] [CrossRef] [Green Version]
- Noothuan, N.; Apitanyasai, K.; Panha, S.; Tassanakajon, A. Snail mucus from the mantle and foot of two land snails, Lissachatina fulica and Hemiplecta distincta, exhibits different protein profile and biological activity. BMC Res. Notes 2021, 14, 138. [Google Scholar] [CrossRef] [PubMed]
- Dolashki, A.; Velkova, L.; Daskalova, E.; Zheleva, N.; Topalova, Y.; Atanasov, V.; Voelter, W.; Dolashka, P. Antimicrobial Activities of Different Fractions from Mucus of the Garden Snail Cornu aspersum. Biomedicines 2020, 8, 315. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, N.G.; Simova, S.D.; Dangalov, M.; Velkova, L.; Atanasov, V.; Dolashki, A.; Dolashka, P. An 1H NMR- and MS-Based Study of Metabolites Profiling of Garden Snail Helix aspersa Mucus. Metabolites 2020, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Velkova, L.; Nissimova, A.; Dolashki, A.; Daskalova, E.; Dolashka, P.; Topalova, Y. Glycine-rich peptides from C. aspersum snail with antibacterial activity. Bulg. Chem. Commun. 2018, 50, 169–175. [Google Scholar]
- Dolashki, A.; Nissimova, A.; Daskalova, E.; Velkova, L.; Topalova, Y.; Hristova, P.; Traldi, P.; Voelter, W.; Dolashka, P. Structure and antibacterial activity of isolated peptides from the mucus of garden snail Cornu aspersum. Bulg. Chem. Commun. C 2018, 50, 195–200. [Google Scholar]
- Meher, P.; Sahu, T.; Saini, V.; Rao, A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, 42362. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Ordóñez, A.; Begley, M.; Clifford, T.; Deasy, T.; Considine, K.; Hill, C. Structure-Activity Relationship of Synthetic Variants of the Milk-Derived Antimicrobial Peptide αs2-Casein f (183–207). Appl. Environ. Microbiol. 2013, 79, 5179–5185. [Google Scholar] [CrossRef] [Green Version]
- Nickel, J.C.; Ruseska, I.; Wright, J.B.; Costerton, J.W. 1985 Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 1985, 27, 619–624. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 2006, 306, 251–258. [Google Scholar]
- Joo, H.S.; Otto, M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 2012, 19, 1503–1513. [Google Scholar] [CrossRef] [Green Version]
- Di Luca, M.; Maccari, G.; Nifosi, R. Treatment of microbial biofilms in the post-antibiotic era: Prophylactic and therapeutic use of antimicrobial peptides and their design by bioinformatics tools. Pathog. Dis. 2014, 70, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strempel, N.; Strehmel, J.; Overhage, J. Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr. Pharm. Des. 2015, 21, 67–84. [Google Scholar] [CrossRef]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, M.; Chiriac, A.I.; Otto, A.; Zweytick, D.; May, C.; Schumacher, C.; Gustg, R.; Albadah, H.B.; Penkovah, M.; Krämeri, U.; et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E1409–E1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, P.; Li, Y.; Cai, J.; Song, L. Selective membrane disruption mechanism of an antibacterial γ-AApeptide defned by EPR spectroscopy. Biophys. J. 2016, 110, 1789–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [Green Version]
- Batoni, G.; Maisetta, G.; Brancatisano, F.L.; Esin, S.; Campa, M. Use of antimicrobial peptides against microbial biofilms: Advantages and limits. Curr. Med. Chem. 2011, 18, 256–279. [Google Scholar] [CrossRef]
- Band, V.I.; Weiss, D.S. Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics 2015, 4, 18–41. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, J.; Yu, W.; Wang, Z.; Li, J.; Feng, X.; Wang, J.; Shan, A. De novo design of a pH-triggered self-assembled β-hairpin nanopeptide with the dual biological functions for antibacterial and entrapment. J. Nanobiotechnol. 2021, 19, 183. [Google Scholar] [CrossRef]
- Sousa, J.C.; Berto, R.F.; Gois, E.A.; Fontenele-Cardi, N.C.; Honório-Júnior, J.E.; Konno, K.; Richardson, M.; Rocha, M.F.; Camargo, A.A.; Pimenta, D.C.; et al. Leptoglycin: A new Glycine/Leucine-rich antimicrobial peptide isolated from the skin secretion of the South American frog Leptodactylus pentadactylus (Leptodactylidae). Toxicon 2009, 54, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opdenakker, G.; Rudd, P.M.; Wormald, M.R.; Dwek, R.A.; Van Damme, J. Cells regulate the activities of cytokines by glycosylation. FASEB J. 1995, 9, 453–457. [Google Scholar] [CrossRef]
- Moradi, S.V.; Hussein, W.M.; Varamini, P.; Simerska, P.; Toth, I. Glycosylation, an effective synthetic strategy to improve the bioavailability of therapeutic peptides. Chem. Sci. 2016, 7, 2492–2500. [Google Scholar] [CrossRef] [Green Version]
- Lele, D.S.; Talat, S.; Kumari, S.; Srivastava, N.; Kaur, K.J. Understanding the importance of glycosylated threonine and stereospecific action of Drosocin, a Proline rich antimicrobial peptide. Eur. J. Med. Chem. 2015, 92, 637–647. [Google Scholar] [CrossRef]
- Bednarska, N.G.; Wren, B.W.; Willcocks, S.J. The importance of the glycosylation of antimicrobial peptides: Natural and synthetic approaches. Drug Discov. Today 2017, 22, 919–926. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Hsu, J.-T.; Chung, P.-H.; Cheng, W.; Jiang, Y.-N.; Ju, Y.-T. Site-specific N-glycosylation of caprine lysostaphin restricts its bacteriolytic activity toward Staphylococcus aureus. Anim. Biotechnol. 2013, 24, 129–147. [Google Scholar] [CrossRef]
- Ilieva, N.; Petkov, P.; Lilkova, E.; Lazarova, T.; Dolashki, A.; Velkova, L.; Dolashka, P.; Litov, L. In Silico Study on the Structure of Novel Natural Bioactive Peptides; Lecture Notes in Computer Science (LNCS) 11958; Springer: Cham, Switzerland, 2020; pp. 332–339. [Google Scholar]
- Trapella, C.; Rizzo, R.; Gallo, S.; Alogna, A.; Bortolotti, D.; Casciano, F.; Zauli, G.; Secchiero, P.; Voltan, R. Helix Complex snail mucus exhibits pro-survival, proliferative and pro-migration effects on mammalian fibroblasts. Sci. Rep. 2018, 8, 17665. [Google Scholar] [CrossRef]
- Li, J.; Shang, L.; Lan, J.; Chou, S.; Feng, X.; Shi, B.; Wang, J.; Lyu, Y.; Shan, A. Targeted and intracellular antibacterial activity against S. agalactiae of the chimeric peptides based on pheromone and cell-penetrating peptides. ACS Appl. Mater. Interfaces 2020, 12, 44459–44474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Shan, A.; Feng, X. Avian host defense cathelicidins: Structure, expression, biological functions, and potential therapeutic applications. Poult. Sci. 2020, 99, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Jiale, Z.; Jian, J.; Xinyi, T.; Haoji, X.; Xueqin, H.; Xiao, W. Design of a novel antimicrobial peptide 1018M targeted ppGpp to inhibit MRSA biofilm formation. AMB Express 2021, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Wang, X.; Preston, J.F., III; Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef] [Green Version]
- Spinosa, M.R.; Progida, C.; Tala, A.; Cogli, L.; Alifano, P.; Bucci, C. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 2007, 75, 3594–3603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onime, L.A.; Oyama, L.B.; Thomas, B.J.; Gani, J.; Alexander, P.; Waddams, K.E.; Cookson, A.; Fernandez-Fuentes, N.; Creevey, C.J.; Huws, S.A. The rumen eukaryotome is a source of novel antimicrobial peptides with therapeutic potential. BMC Microbiol. 2021, 21, 105. [Google Scholar] [CrossRef] [PubMed]












| No. | Amino Acid Sequence of Peptides (N-C-Terminals), De Novo Sequencing. | MALDI [M + H]+ (Da) | Calc. Mass (Da) | pI | GRAVY | Net Charge | Predict. Antibacterial (%) | Predict. Antiviral (%) | Predict. Antifungal (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AAGLAGAGGGGGG | 872.42 | 871.41 | 5.57 | 0.600 | 0/0 | 58 | 41 | 32 |
| 2 | DKGLGGFEA | 893.51 | 892.43 | 4.37 | −0.411 | −2/+1 | 30 | 29 | 20 |
| 3 | LGDLNAEFAAG | 1077.67 | 1076.51 | 3.67 | 0.409 | −2/0 | 27 | 46 | 30 |
| 4 | AGVGAGGANPSTYVG | 1277.91 | 1276.60 | 5.57 | 0.260 | 0/0 | 25 | 7.5 | 11 |
| 5 | GAACNLEDGSCLGV | 1308.81 | 1307.55 | 3.67 | 0.564 | −2/0 | 58 | 58 | 53 |
| 6 | EPGGGGGEGGGLLGVAL | 1396.96 | 1395.70 | 3.80 | 0.306 | −2/0 | 41 | 35 | 20 |
| 7 | LGPLYDEMGPVGGDVG | 1576.74 | 1574.73 | 3.49 | 0.056 | −3/0 | 9.7 | 20 | 6.2 |
| 8 | ASKGCGPGSCPPGDTVAGVG | 1716.82 | 1715.76 | 5.86 | 0.005 | −1/+1 | 25 | 12 | 23 |
| 9 | ACSLLLGGGGVGGGKGGGGHAG | 1739.02 | 1737.86 | 8.27 | 0.409 | 0/+1 | 83 | 49 | 67 |
| 10 | ACLTPVDHFFAGMPCGGGP | 1877.14 | 1875.81 | 5.08 | 0.542 | −1/0 | 32 | 43 | 20 |
| 11 | NGLFGGLGGGGHGGGGKGPGEGGG | 1909.90 | 1908.88 | 6.75 | −0.487 | −1/+1 | 90 | 67 | 80 |
| 12 | LLLLMLGGGLVGGLLGGGGKGGG | 1966.24 | 1965.14 | 8.75 | 1.209 | 0/+1 | 92 | 57 | 76 |
| 13 | PFLLGVGGLLGGSVGGGGGGGGAPL | 2023.14 | 2022.09 | 5.96 | 0.912 | 0/0 | 69 | 32 | 38 |
| 14 | GMVVKHCSAPLDSFAEFAGA | 2036.93 | 2035.95 | 5.32 | 0.565 | −2/+1 | 42 | 20 | 26 |
| 15 * | GLLGGGGGAGGGGLVGGLLNG | 1609.94 | 1608.86 | 5.52 | 0.776 | 0/+1 | 90.0 | 53.6 | 65.0 |
| 16 * | MGGLLGGVNGGGKGGGGPGAP | 1666.83 | 1665.83 | 8.5 | 0.005 | 0/+1 | 78.6 | 52.0 | 61.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topalova, Y.; Belouhova, M.; Velkova, L.; Dolashki, A.; Zheleva, N.; Daskalova, E.; Kaynarov, D.; Voelter, W.; Dolashka, P. Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785. Biomedicines 2022, 10, 672. https://doi.org/10.3390/biomedicines10030672
Topalova Y, Belouhova M, Velkova L, Dolashki A, Zheleva N, Daskalova E, Kaynarov D, Voelter W, Dolashka P. Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785. Biomedicines. 2022; 10(3):672. https://doi.org/10.3390/biomedicines10030672
Chicago/Turabian StyleTopalova, Yana, Mihaela Belouhova, Lyudmila Velkova, Aleksandar Dolashki, Nellie Zheleva, Elmira Daskalova, Dimitar Kaynarov, Wolfgang Voelter, and Pavlina Dolashka. 2022. "Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785" Biomedicines 10, no. 3: 672. https://doi.org/10.3390/biomedicines10030672
APA StyleTopalova, Y., Belouhova, M., Velkova, L., Dolashki, A., Zheleva, N., Daskalova, E., Kaynarov, D., Voelter, W., & Dolashka, P. (2022). Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785. Biomedicines, 10(3), 672. https://doi.org/10.3390/biomedicines10030672