A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy and Information Sources
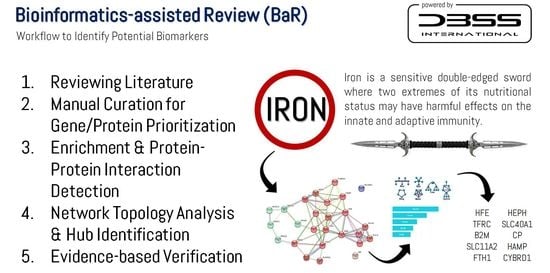
2.2. Manual Curation and Bioinformatics-Assisted Review
2.3. Identification of Potential Biomarkers
3. Iron Uptake and Metabolism
4. Iron and the Immune System
5. Identification of Potential Biomarkers of Stress-Induced Immunosuppression
5.1. Evidence-Based Verification of the Identified Potential Biomarkers
5.2. Limitations, Strengths, and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Wu, Y.; Li, T.; Wang, X.; Zhu, C. Iron Metabolism and Brain Development in Premature Infants. Front. Physiol. 2019, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.; Parish, H.; Smith, A.; Vella, F.; Stirling, J. Oxford Dictionary of Biochemistry and Molecular Biology; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Ciaccio, C.; Coletta, A.; Coletta, M. Role of hemoglobin structural-functional relationships in oxygen transport. Mol. Asp. Med. 2021, 84, 101022. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.; Pfeiffer, C.M.; Georgieff, M.K.; Brittenham, G.; Fairweather-Tait, S.; Hurrell, R.F.; McArdle, H.J.; Raiten, D.J. Biomarkers of Nutrition for Development (BOND)—Iron Review. J. Nutr. 2018, 148, 1001S–1067S. [Google Scholar] [CrossRef] [Green Version]
- Silva, B.; Faustino, P. An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim. Biophys. Acta—Mol. Basis Dis. 2015, 1852, 1347–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, I.; Parker, H.M.; Rangan, A.; Prvan, T.; Cook, R.L.; Donges, C.E.; Steinbeck, K.S.; O’Dwyer, N.J.; Cheng, H.L.; Franklin, J.L.; et al. Association between Haem and Non-Haem Iron Intake and Serum Ferritin in Healthy Young Women. Nutrients 2018, 10, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Institute of Medicine (US) Panel on Micronutrients. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Kennedy, E.; Meyers, L. Dietary Reference Intakes: Development and uses for assessment of micronutrient status of women—A global perspective. Am. J. Clin. Nutr. 2005, 81, 1194S–1197S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenkin, A. Dietary reference values for vitamin A, vitamin K, arsenic, boron, chromium, copper, iodine, iron, manganese, molybdenum, nickel, silicon, vanadium and zinc. J. Hum. Nutr. Diet. 2003, 16, 199–200. [Google Scholar] [CrossRef]
- Hurrell, R.; Egli, I. Iron bioavailability and dietary reference values. Am. J. Clin. Nutr. 2010, 91, 1461S–1467S. [Google Scholar] [CrossRef]
- Blanco-Rojo, R.; Vaquero, M.P. Iron bioavailability from food fortification to precision nutrition. A review. Innov. Food Sci. Emerg. Technol. 2019, 51, 126–138. [Google Scholar] [CrossRef]
- Yiannikourides, A.; Latunde-Dada, G. A Short Review of Iron Metabolism and Pathophysiology of Iron Disorders. Medicines 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.F. The Regulation of Iron Absorption and Homeostasis. Clin. Biochemist. Rev. 2016, 37, 51–62. [Google Scholar]
- Vogt, A.S.; Arsiwala, T.; Mohsen, M.; Vogel, M.; Manolova, V.; Bachmann, M.F. On Iron Metabolism and Its Regulation. Int. J. Mol. Sci. 2021, 22, 4591. [Google Scholar] [CrossRef] [PubMed]
- Green, R.; Charlton, R.; Seftel, H.; Bothwell, T.; Mayet, F.; Adams, B.; Finch, C.; Layrisse, M. Body iron excretion in man: A collaborative study. Am. J. Med. 1968, 45, 336–353. [Google Scholar] [CrossRef]
- Hunt, J.R.; Zito, C.A.; Johnson, L.K. Body iron excretion by healthy men and women. Am. J. Clin. Nutr. 2009, 89, 1792–1798. [Google Scholar] [CrossRef] [Green Version]
- Mercadante, C.J.; Prajapati, M.; Parmar, J.H.; Conboy, H.L.; Dash, M.E.; Pettiglio, M.A.; Herrera, C.; Bu, J.T.; Stopa, E.G.; Mendes, P.; et al. Gastrointestinal iron excretion and reversal of iron excess in a mouse model of inherited iron excess. Haematologica 2019, 104, 678–689. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Perez-Idarraga, A.; Odriozola-Martinez, A.; Kreider, R.B. The 4R’s Framework of Nutritional Strategies for Post-Exercise Recovery: A Review with Emphasis on New Generation of Carbohydrates. Int. J. Environ. Res. Public Health 2020, 18, 103. [Google Scholar] [CrossRef]
- Selye, H. A Syndrome produced by Diverse Nocuous Agents. Nature 1936, 138, 32. [Google Scholar] [CrossRef]
- Zhang, Q.; Bhattacharya, S.; Andersen, M.E.; Conolly, R.B. Computational Systems Biology and Dose-Response Modeling in Relation to New Directions in Toxicity Testing. J. Toxicol. Environ. Health Part B 2010, 13, 253–276. [Google Scholar] [CrossRef]
- Zhang, Q.; Bhattacharya, S.; Pi, J.; Clewell, R.A.; Carmichael, P.L.; Andersen, M.E. Adaptive Posttranslational Control in Cellular Stress Response Pathways and Its Relationship to Toxicity Testing and Safety Assessment. Toxicol. Sci. 2015, 147, 302–316. [Google Scholar] [CrossRef] [Green Version]
- Kapuy, O.; Márton, M.; Bánhegyi, G.; Vinod, P.K. Multiple system-level feedback loops control life-and-death decisions in endoplasmic reticulum stress. FEBS Lett. 2020, 594, 1112–1123. [Google Scholar] [CrossRef] [Green Version]
- Sterling, P.; Eyer, J. Biological basis of stress-related mortality. Soc. Sci. Med. E 1981, 15, 3–42. [Google Scholar] [CrossRef]
- Sterling, P.; Eyer, J. Allostasis: A new paradigm to explain arousal pathology. In Handbook of Life Stress, Cognition and Health; John Wiley & Sons: Oxford, UK, 1988; pp. 629–649. [Google Scholar]
- McEwen, B.S. Stress, Adaptation, and Disease: Allostasis and Allostatic Load. Ann. N. Y. Acad. Sci. 1998, 840, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Sterling, P. Allostasis: A model of predictive regulation. Physiol. Behav. 2012, 106, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Guidi, J.; Lucente, M.; Sonino, N.; Fava, G.A. Allostatic Load and Its Impact on Health: A Systematic Review. Psychother. Psychosom. 2021, 90, 11–27. [Google Scholar] [CrossRef]
- Dhabhar, F.S. Effects of stress on immune function: The good, the bad, and the beautiful. Immunol. Res. 2014, 58, 193–210. [Google Scholar] [CrossRef]
- Martin, L.B. Stress and immunity in wild vertebrates: Timing is everything. Gen. Comp. Endocrinol. 2009, 163, 70–76. [Google Scholar] [CrossRef]
- Justiz Vaillant, A.A.; Qurie, A. Immunodeficiency; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Ruini, C.; Offidani, E.; Vescovelli, F. Life Stressors, Allostatic Overload, and Their Impact on Posttraumatic Growth. J. Loss Trauma 2014, 20, 109–122. [Google Scholar] [CrossRef]
- Mastorakos, G.; Pavlatou, M. Exercise as a stress model and the interplay between the hypothalamus-pituitary-adrenal and the hypothalamus-pituitary-thyroid axes. Horm. Metab. Res. 2005, 37, 577–584. [Google Scholar] [CrossRef]
- Manolis, A.S.; Manolis, A.A. Exercise and Arrhythmias: A Double-Edged Sword. Pacing Clin. Electrophysiol. 2016, 39, 748–762. [Google Scholar] [CrossRef]
- Luscher, T.F. Sport, exercise, and daily activity: A double-edged sword revisited. Eur. Heart J. 2016, 37, 2505–2507. [Google Scholar] [CrossRef] [Green Version]
- D’Alessio, L.; Korman, G.P.; Sarudiansky, M.; Guelman, L.R.; Scevola, L.; Pastore, A.; Obregon, A.; Roldan, E.J.A. Reducing Allostatic Load in Depression and Anxiety Disorders: Physical Activity and Yoga Practice as Add-On Therapies. Front. Psychiatry 2020, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Reichel, T.; Zeilinger, C. Role of heat shock proteins 70/90 in exercise physiology and exercise immunology and their diagnostic potential in sports. J. Appl. Physiol. (1985) 2019, 126, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Nairz, M.; Haschka, D.; Demetz, E.; Weiss, G. Iron at the interface of immunity and infection. Front. Pharmacol. 2014, 5, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaban, E.P.; Cox, J.V.; Snell, P.; Vaughan, R.H.; Frenkel, E.P. The frequency of anemia and iron deficiency in the runner. Med. Sci. Sports Exerc. 1989, 21, 643–648. [Google Scholar] [CrossRef]
- Schumacher, Y.O.; Schmid, A.; Grathwohl, D.; Bültermann, D.; Berg, A. Hematological indices and iron status in athletes of various sports and performances. Med. Sci. Sports Exerc. 2002, 34, 869–875. [Google Scholar] [CrossRef]
- Wouthuyzen-Bakker, M.; van Assen, S. Exercise-induced anaemia: A forgotten cause of iron deficiency anaemia in young adults. Br. J. Gen. Pract. 2015, 65, 268. [Google Scholar] [CrossRef] [Green Version]
- Mountjoy, M.; Sundgot-Borgen, J.K.; Burke, L.M.; Ackerman, K.E.; Blauwet, C.; Constantini, N.; Lebrun, C.; Lundy, B.; Melin, A.K.; Meyer, N.L.; et al. IOC consensus statement on relative energy deficiency in sport (RED-S): 2018 update. Br. J. Sports Med. 2018, 52, 687–697. [Google Scholar] [CrossRef] [Green Version]
- Damian, M.-T.; Vulturar, R.; Login, C.C.; Damian, L.; Chis, A.; Bojan, A. Anemia in Sports: A Narrative Review. Life 2021, 11, 987. [Google Scholar] [CrossRef]
- Akil, M.; Celenk, C. Iron metabolism and importance of iron in exercise. Int. J. Acad. Res. 2013, 5, 223–230. [Google Scholar] [CrossRef]
- Burke, L.M.; Close, G.L.; Lundy, B.; Mooses, M.; Morton, J.P.; Tenforde, A.S. Relative Energy Deficiency in Sport in Male Athletes: A Commentary on Its Presentation Among Selected Groups of Male Athletes. Int. J. Sport Nutr. Exerc. Metab. 2018, 28, 364–374. [Google Scholar] [CrossRef]
- Castell, L.M.; Nieman, D.C.; Bermon, S.; Peeling, P. Exercise-Induced Illness and Inflammation: Can Immunonutrition and Iron Help? Int. J. Sport Nutr. Exerc. Metab. 2019, 29, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.-N.; Gao, G.; Chang, Y.-Z. Hepcidin and sports anemia. Cell Biosci. 2014, 4, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, M.; Garvican-Lewis, L.A.; Cox, G.R.; Govus, A.; McKay, A.K.A.; Stellingwerff, T.; Peeling, P. Iron considerations for the athlete: A narrative review. Eur. J. Appl. Physiol. 2019, 119, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P.; et al. Biomarkers in Stress Related Diseases/Disorders: Diagnostic, Prognostic, and Therapeutic Values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- McKay, A.K.A.; Pyne, D.B.; Burke, L.M.; Peeling, P. Iron Metabolism: Interactions with Energy and Carbohydrate Availability. Nutrients 2020, 12, 3692. [Google Scholar] [CrossRef] [PubMed]
- Valdez, G.; Koob, G. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: Implications for the development of alcoholism. Pharmacol. Biochem. Behav. 2004, 79, 671–689. [Google Scholar] [CrossRef]
- Juster, R.-P.; Seeman, T.; McEwen, B.S.; Picard, M.; Mahar, I.; Mechawar, N.; Sindi, S.; Smith, N.G.; Souza-Talarico, J.; Sarnyai, Z.; et al. Social Inequalities and the Road to Allostatic Load: From Vulnerability to Resilience. In Developmental Psychopathology; Wiley: New York, NY, USA, 2016; pp. 1–54. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Moreno, Y.; Gho, C.; Petro, J.L.; Odriozola-Martinez, A.; Kreider, R.B. Effects of Ashwagandha (Withania somnifera) on Physical Performance: Systematic Review and Bayesian Meta-Analysis. J. Funct. Morphol. Kinesiol. 2021, 6, 20. [Google Scholar] [CrossRef]
- Whittemore, R.; Knafl, K. The integrative review: Updated methodology. J. Adv. Nurs. 2005, 52, 546–553. [Google Scholar] [CrossRef]
- Hopia, H.; Latvala, E.; Liimatainen, L. Reviewing the methodology of an integrative review. Scand. J. Caring Sci. 2016, 30, 662–669. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Kreider, R.B.; Stout, J.R.; Forero, D.A.; Kerksick, C.M.; Roberts, M.D.; Rawson, E.S. Metabolic Basis of Creatine in Health and Disease: A Bioinformatics-Assisted Review. Nutrients 2021, 13, 1238. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Moreno, Y.; Rawson, E.S.; Forero, D.A.; Stout, J.R.; Kerksick, C.M.; Roberts, M.D.; Kreider, R.B. A Convergent Functional Genomics Analysis to Identify Biological Regulators Mediating Effects of Creatine Supplementation. Nutrients 2021, 13, 2521. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Ma’ayan, A. KEA: Kinase enrichment analysis. Bioinformatics 2009, 25, 684–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Csermely, P.; Tian, K.; Rajendran, R.; Doddananjaiah, M.; Krstic-Demonacos, M.; Schwartz, J.-M. Dynamics of DNA Damage Induced Pathways to Cancer. PLoS ONE 2013, 8, e72303. [Google Scholar] [CrossRef] [Green Version]
- Rzhetsky, A.; Bakker, E.; Tian, K.; Mutti, L.; Demonacos, C.; Schwartz, J.-M.; Krstic-Demonacos, M. Insight into glucocorticoid receptor signalling through interactome model analysis. PLoS Comput. Biol. 2017, 13, e1005825. [Google Scholar] [CrossRef] [Green Version]
- Theodosiou, T.; Efstathiou, G.; Papanikolaou, N.; Kyrpides, N.C.; Bagos, P.G.; Iliopoulos, I.; Pavlopoulos, G.A. NAP: The Network Analysis Profiler, a web tool for easier topological analysis and comparison of medium-scale biological networks. BMC Res. Notes 2017, 10, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolotareva, O.; Kleine, M. A Survey of Gene Prioritization Tools for Mendelian and Complex Human Diseases. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Odell, S.G.; Lazo, G.R.; Woodhouse, M.R.; Hane, D.L.; Sen, T.Z. The art of curation at a biological database: Principles and application. Curr. Plant Biol. 2017, 11–12, 2–11. [Google Scholar] [CrossRef]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef]
- Ortutay, C.; Vihinen, M. Immunome knowledge base (IKB): An integrated service for immunome research. BMC Immunol. 2009, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Bartok, B.; Oler, E.; Liang, K.Y.H.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An online database of molecular biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Systemic Iron Homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latunde-Dada, G.O.; Van der Westhuizen, J.; Vulpe, C.D.; Anderson, G.J.; Simpson, R.J.; McKie, A.T. Molecular and Functional Roles of Duodenal Cytochrome B (Dcytb) in Iron Metabolism. Blood Cells Mol. Dis. 2002, 29, 356–360. [Google Scholar] [CrossRef]
- Gulec, S.; Anderson, G.J.; Collins, J.F. Mechanistic and regulatory aspects of intestinal iron absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G397–G409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wißbrock, A.; Paul George, A.A.; Brewitz, H.H.; Kühl, T.; Imhof, D. The molecular basis of transient heme-protein interactions: Analysis, concept and implementation. Biosci. Rep. 2019, 39, 39. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel-Abramowski, S.; Waeber, G.; Gassner, C.; Buser, A.; Frey, B.M.; Favrat, B.; Tissot, J.D. Physiology of iron metabolism. Transfus. Med. Hemotherapy Off. Organ Der Dtsch. Ges. Transfus. Immunhamatol. 2014, 41, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Graversen, J.H.; Madsen, M.; Moestrup, S.K. CD163: A signal receptor scavenging haptoglobin-hemoglobin complexes from plasma. Int. J. Biochem. Cell Biol. 2002, 34, 309–314. [Google Scholar] [CrossRef]
- Latunde-Dada, G.O.; Takeuchi, K.; Simpson, R.J.; McKie, A.T. Haem carrier protein 1 (HCP1): Expression and functional studies in cultured cells. FEBS Lett. 2006, 580, 6865–6870. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Y.; Paragas, N.; Ned, R.M.; Qiu, A.; Viltard, M.; Leete, T.; Drexler, I.R.; Chen, X.; Sanna-Cherchi, S.; Mohammed, F.; et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev. Cell 2009, 16, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Arosio, P.; Elia, L.; Poli, M. Ferritin, cellular iron storage and regulation. IUBMB Life 2017, 69, 414–422. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin: The Protein Nanocage and Iron Biomineral in Health and in Disease. Inorg. Chem. 2013, 52, 12223–12233. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.J.; Frazer, D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017, 106, 1559s–1566s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philpott, C.C.; Ryu, M.S.; Frey, A.; Patel, S. Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J. Biol. Chem. 2017, 292, 12764–12771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drakesmith, H.; Nemeth, E.; Ganz, T. Ironing out Ferroportin. Cell Metab. 2015, 22, 777–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, D.R.; Lane, D.J.R.; Becker, E.M.; Huang, M.L.H.; Whitnall, M.; Rahmanto, Y.S.; Sheftel, A.D.; Ponka, P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. USA 2010, 107, 10775. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Walden, W.E.; Theil, E.C.; Goss, D.J. Thermodynamic and Kinetic Analyses of Iron Response Element (IRE)-mRNA Binding to Iron Regulatory Protein, IRP1. Sci. Rep. 2017, 7, 8532. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 2014, 5, 176. [Google Scholar] [CrossRef] [Green Version]
- Cronin, S.J.F.; Woolf, C.J.; Weiss, G.; Penninger, J.M. The Role of Iron Regulation in Immunometabolism and Immune-Related Disease. Front. Mol. Biosci. 2019, 6, 116. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.D.; Tan, E.K. Iron regulatory protein (IRP)-iron responsive element (IRE) signaling pathway in human neurodegenerative diseases. Mol. Neurodegener. 2017, 12, 75. [Google Scholar] [CrossRef]
- Chen, S.-C.; Olsthoorn, R.C.L. Relevance of the iron-responsive element (IRE) pseudotriloop structure for IRP1/2 binding and validation of IRE-like structures using the yeast three-hybrid system. Gene 2019, 710, 399–405. [Google Scholar] [CrossRef]
- Vulpe, C.D.; Kuo, Y.M.; Murphy, T.L.; Cowley, L.; Askwith, C.; Libina, N.; Gitschier, J.; Anderson, G.J. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 1999, 21, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Petrak, J.; Vyoral, D. Hephaestin—A ferroxidase of cellular iron export. Int. J. Biochem. Cell Biol. 2005, 37, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, V.B. Looking for a partner: Ceruloplasmin in protein-protein interactions. Biometals 2019, 32, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allden, S.J.; Ogger, P.P.; Ghai, P.; McErlean, P.; Hewitt, R.; Toshner, R.; Walker, S.A.; Saunders, P.; Kingston, S.; Molyneaux, P.L.; et al. The Transferrin Receptor CD71 Delineates Functionally Distinct Airway Macrophage Subsets during Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 200, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Otaki, Y.; Nakanishi, T.; Hasuike, Y.; Moriguchi, R.; Nanami, M.; Hama, Y.; Izumi, M.; Takamitsu, Y. Defective regulation of iron transporters leading to iron excess in the polymorphonuclear leukocytes of patients on maintenance hemodialysis. Am. J. Kidney Dis. 2004, 43, 1030–1039. [Google Scholar] [CrossRef]
- Maneva, A.; Taleva, B. Receptors for Transferrin on Human Neutrophils. Biotechnol. Biotechnol. Equip. 2009, 23, 477–479. [Google Scholar] [CrossRef]
- Larrick, J.W.; Cresswell, P. Modulation of cell surface iron transferrin receptors by cellular density and state of activation. J. Supramol. Struct. 1979, 11, 579–586. [Google Scholar] [CrossRef]
- Neckers, L.M.; Cossman, J. Transferrin receptor induction in mitogen-stimulated human T lymphocytes is required for DNA synthesis and cell division and is regulated by interleukin 2. Proc. Natl. Acad. Sci. USA 1983, 80, 3494. [Google Scholar] [CrossRef] [Green Version]
- Schweier, O.; Hofmann, M.; Pircher, H. KLRG1 activity is regulated by association with the transferrin receptor. Eur. J. Immunol. 2014, 44, 1851–1856. [Google Scholar] [CrossRef]
- Sottile, R.; Federico, G.; Garofalo, C.; Tallerico, R.; Faniello, M.C.; Quaresima, B.; Cristiani, C.M.; Di Sanzo, M.; Cuda, G.; Ventura, V.; et al. Iron and Ferritin Modulate MHC Class I Expression and NK Cell Recognition. Front. Immunol. 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neckers, L.M.; Yenokida, G.; James, S.P. The role of the transferrin receptor in human B lymphocyte activation. J. Immunol. 1984, 133, 2437–2441. [Google Scholar] [PubMed]
- Hamilton, T.A.; Weiel, J.E.; Adams, D.O. Expression of the transferrin receptor in murine peritoneal macrophages is modulated in the different stages of activation. J. Immunol. 1984, 132, 2285–2290. [Google Scholar] [PubMed]
- Corna, G.; Caserta, I.; Monno, A.; Apostoli, P.; Manfredi, A.A.; Camaschella, C.; Rovere-Querini, P. The Repair of Skeletal Muscle Requires Iron Recycling through Macrophage Ferroportin. J. Immunol. 2016, 197, 1914. [Google Scholar] [CrossRef] [Green Version]
- Gammella, E.; Buratti, P.; Cairo, G.; Recalcati, S. Macrophages: Central regulators of iron balance. Metallomics 2014, 6, 1336–1345. [Google Scholar] [CrossRef] [Green Version]
- Maio, N.; Zhang, D.L.; Ghosh, M.C.; Jain, A.; SantaMaria, A.M.; Rouault, T.A. Mechanisms of cellular iron sensing, regulation of erythropoiesis and mitochondrial iron utilization. Semin. Hematol. 2021, 58, 161–174. [Google Scholar] [CrossRef]
- Bowlus, C.L. The role of iron in T cell development and autoimmunity. Autoimmun. Rev. 2003, 2, 73–78. [Google Scholar] [CrossRef]
- Mu, Q.; Chen, L.; Gao, X.; Shen, S.; Sheng, W.; Min, J.; Wang, F. The role of iron homeostasis in remodeling immune function and regulating inflammatory disease. Sci. Bull. 2021, 66, 1806–1816. [Google Scholar] [CrossRef]
- Martins, A.C.; Almeida, J.I.; Lima, I.S.; Kapitao, A.S.; Gozzelino, R. Iron Metabolism and the Inflammatory Response. IUBMB Life 2017, 69, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Nappi, A.J.; Vass, E. Iron, metalloenzymes and cytotoxic reactions. Cell Mol. Biol. (Noisy-Le-Grand) 2000, 46, 637–647. [Google Scholar]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Tsaousis, A.D. On the Origin of Iron/Sulfur Cluster Biosynthesis in Eukaryotes. Front. Microbiol. 2019, 10, 2478. [Google Scholar] [CrossRef] [PubMed]
- Pantopoulos, K.; Hentze, M.W. Activation of iron regulatory protein-1 by oxidative stress in vitro. Proc. Natl. Acad. Sci. USA 1998, 95, 10559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yauger, Y.J.; Bermudez, S.; Moritz, K.E.; Glaser, E.; Stoica, B.; Byrnes, K.R. Iron accentuated reactive oxygen species release by NADPH oxidase in activated microglia contributes to oxidative stress in vitro. J. Neuroinflamm. 2019, 16, 41. [Google Scholar] [CrossRef]
- Paino, I.M.M.; Miranda, J.C.; Marzocchi-Machado, C.M.; Cesarino, E.J.; de Castro, F.A.; de Souza, A.M. Phagocytosis, Oxidative Burst, and Produced Reactive Species are Affected by Iron Deficiency Anemia and Anemia of Chronic Diseases in Elderly. Biol. Trace Elem. Res. 2009, 129, 116–125. [Google Scholar] [CrossRef]
- Nairz, M.; Theurl, I.; Swirski, F.K.; Weiss, G. “Pumping iron”—How macrophages handle iron at the systemic, microenvironmental, and cellular levels. Pflug. Arch. Eur. J. Physiol. 2017, 469, 397–418. [Google Scholar] [CrossRef] [Green Version]
- Wessling-Resnick, M. Iron homeostasis and the inflammatory response. Annu. Rev. Nutr. 2010, 30, 105–122. [Google Scholar] [CrossRef] [Green Version]
- Byrd, T.F.; Horwitz, M.A. Regulation of transferrin receptor expression and ferritin content in human mononuclear phagocytes. Coordinate upregulation by iron transferrin and downregulation by interferon gamma. J. Clin. Investig. 1993, 91, 969–976. [Google Scholar] [CrossRef] [Green Version]
- Hognon, C.; Bignon, E.; Harle, G.; Touche, N.; Grandemange, S.; Monari, A. The Iron Maiden. Cytosolic Aconitase/IRP1 Conformational Transition in the Regulation of Ferritin Translation and Iron Hemostasis. Biomolecules 2021, 11, 1329. [Google Scholar] [CrossRef]
- Kernan, K.F.; Carcillo, J.A. Hyperferritinemia and inflammation. Int. Immunol. 2017, 29, 401–409. [Google Scholar] [CrossRef]
- Hassan, T.H.; Badr, M.A.; Karam, N.A.; Zkaria, M.; El Saadany, H.F.; Abdel Rahman, D.M.; Shahbah, D.A.; Al Morshedy, S.M.; Fathy, M.; Esh, A.M.H.; et al. Impact of iron deficiency anemia on the function of the immune system in children. Medicine 2016, 95, e5395. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.S.; Fayed, H.M.; Ismail, A.M.; Abdel Hakeem, G.L. Assessment of peripheral blood lymphocyte subsets in children with iron deficiency anemia. BMC Pediatr. 2018, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Díaz, V. Regulation of Iron Metabolism and Exercise. Med. Sport. 2011, 15, 230–238. [Google Scholar] [CrossRef]
- Kuvibidila, S.; Baliga, B.S.; Murthy, K.K. Impaired protein kinase C activation as one of the possible mechanisms of reduced lymphocyte proliferation in iron deficiency in mice. Am. J. Clin. Nutr. 1991, 54, 944–950. [Google Scholar] [CrossRef]
- Oexle, H.; Kaser, A.; Möst, J.; Bellmann-Weiler, R.; Werner, E.R.; Werner-Felmayer, G.; Weiss, G. Pathways for the regulation of interferon-γ-inducible genes by iron in human monocytic cells. J. Leukoc. Biol. 2003, 74, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Kuvibidila, S.; Warrier, R.P.; Surendra Baliga, B. An overview of the role of iron in T cell activation. J. Trace Elem. Exp. Med. 2003, 16, 219–225. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Wu, Q.; An, P.; Huang, L.; Wang, J.; Chen, C.; Chen, X.; Zhang, F.; Ma, L.; et al. Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat. Commun. 2019, 10, 2935. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, E.; Ganz, T. The Role of Hepcidin in Iron Metabolism. Acta Haematol. 2009, 122, 78–86. [Google Scholar] [CrossRef] [Green Version]
- Daher, R.; Manceau, H.; Karim, Z. Iron metabolism and the role of the iron-regulating hormone hepcidin in health and disease. Presse Med. (Paris, France: 1983) 2017, 46, e272–e278. [Google Scholar] [CrossRef]
- Zhao, N.; Zhang, A.-S.; Enns, C.A. Iron regulation by hepcidin. J. Clin. Investig. 2013, 123, 2337–2343. [Google Scholar] [CrossRef]
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, A.; Neitz, S.; Mägert, H.J.; Schulz, A.; Forssmann, W.G.; Schulz-Knappe, P.; Adermann, K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000, 480, 147–150. [Google Scholar] [CrossRef] [Green Version]
- Pigeon, C.; Ilyin, G.; Courselaud, B.; Leroyer, P.; Turlin, B.; Brissot, P.; Loréal, O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 2001, 276, 7811–7819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, M.P.; Meynard, D.; Coppin, H. Regulators of hepcidin expression. Vitam. Horm. 2019, 110, 101–129. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Goldberg, Y.P.; Ganz, T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood 2005, 106, 2884–2889. [Google Scholar] [CrossRef]
- De Domenico, I.; Ward, D.M.; Kaplan, J. Hepcidin regulation: Ironing out the details. J. Clin. Investig. 2007, 117, 1755–1758. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef] [Green Version]
- Aschemeyer, S.; Qiao, B.; Stefanova, D.; Valore, E.V.; Sek, A.C.; Ruwe, T.A.; Vieth, K.R.; Jung, G.; Casu, C.; Rivella, S.; et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood 2018, 131, 899–910. [Google Scholar] [CrossRef]
- Cherayil, B.J. Iron and Immunity: Immunological Consequences of Iron Deficiency and Overload. Arch. Immunol. Ther. Exp. 2010, 58, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Ahluwalia, N.; Sun, J.; Krause, D.; Mastro, A.; Handte, G. Immune function is impaired in iron-deficient, homebound, older women. Am. J. Clin. Nutr. 2004, 79, 516–521. [Google Scholar] [CrossRef] [Green Version]
- Barton, J.C.; Acton, R.T. Hepcidin, iron, and bacterial infection. Vitam. Horm. 2019, 110, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Michels, K.; Nemeth, E.; Ganz, T.; Mehrad, B. Hepcidin and Host Defense against Infectious Diseases. PLoS Pathog. 2015, 11, e1004998. [Google Scholar] [CrossRef] [Green Version]
- Girelli, D.; Marchi, G.; Busti, F.; Vianello, A. Iron metabolism in infections: Focus on COVID-19. Semin. Hematol. 2021, 58, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.P.; Turner, J.E. Debunking the Myth of Exercise-Induced Immune Suppression: Redefining the Impact of Exercise on Immunological Health Across the Lifespan. Front. Immunol. 2018, 9, 648. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, R.; Carbone, L.; Petro, J.L.; Cione, E.; Vargas, S.; Angulo, H.; Forero, D.A.; Odriozola-Martinez, A.; Kreider, R.B.; Bonilla, D.A. Sarcopenia: Etiology, Nutritional Approaches, and miRNAs. Int. J. Mol. Sci. 2021, 22, 9724. [Google Scholar] [CrossRef]
- Nieman, D.C.; Wentz, L.M. The compelling link between physical activity and the body’s defense system. J. Sport Health Sci. 2019, 8, 201–217. [Google Scholar] [CrossRef]
- Suzuki, K.; Hayashida, H. Effect of Exercise Intensity on Cell-Mediated Immunity. Sports 2021, 9, 8. [Google Scholar] [CrossRef]
- Xu, G.; Lin, W.; McAinch, A.J.; Yan, X.; Weng, X. Identification of Urinary Biomarkers for Exercise-Induced Immunosuppression by iTRAQ Proteomics. BioMed Res. Int. 2020, 2020, 3030793. [Google Scholar] [CrossRef] [Green Version]
- Estruel-Amades, S.; Camps-Bossacoma, M.; Massot-Cladera, M.; Perez-Cano, F.J.; Castell, M. Alterations in the innate immune system due to exhausting exercise in intensively trained rats. Sci. Rep. 2020, 10, 967. [Google Scholar] [CrossRef]
- Scheffer, D.D.L.; Latini, A. Exercise-induced immune system response: Anti-inflammatory status on peripheral and central organs. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165823. [Google Scholar] [CrossRef]
- Nieman, D.C.; Pence, B.D. Exercise immunology: Future directions. J. Sport Health Sci. 2020, 9, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Oldham, S.; Fulcher, B.; Parkes, L.; Arnatkevic Iute, A.; Suo, C.; Fornito, A. Consistency and differences between centrality measures across distinct classes of networks. PLoS ONE 2019, 14, e0220061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.C.; Fragala, M.S.; Kavouras, S.A.; Queen, R.M.; Pryor, J.L.; Casa, D.J. Biomarkers in Sports and Exercise: Tracking Health, Performance, and Recovery in Athletes. J. Strength Cond. Res. 2017, 31, 2920–2937. [Google Scholar] [CrossRef] [Green Version]
- Chicharro, J.L.; Hoyos, J.; Gomez-Gallego, F.; Villa, J.G.; Bandres, F.; Celaya, P.; Jimenez, F.; Alonso, J.M.; Cordova, A.; Lucia, A. Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br. J. Sports Med. 2004, 38, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.C.; Edwards, C.Q.; Acton, R.T. HFE gene: Structure, function, mutations, and associated iron abnormalities. Gene 2015, 574, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Tangudu, N.K.; Yilmaz, D.; Worle, K.; Gruber, A.; Colucci, S.; Leopold, K.; Muckenthaler, M.U.; Vujic Spasic, M. Macrophage-HFE controls iron metabolism and immune responses in aged mice. Haematologica 2021, 106, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Luszczyk, M.; Kaczorowska-Hac, B.; Milosz, E.; Adamkiewicz-Drozynska, E.; Ziemann, E.; Laskowski, R.; Flis, D.; Rokicka-Hebel, M.; Antosiewicz, J. Reduction of Skeletal Muscle Power in Adolescent Males Carrying H63D Mutation in the HFE Gene. BioMed Res. Int. 2017, 2017, 5313914. [Google Scholar] [CrossRef] [Green Version]
- Kaczorowska-Hac, B.; Luszczyk, M.; Antosiewicz, J.; Kaczor, J.J. Iron metabolism and hepcidin concentration in teenagers before and after exercise in relation to the HFE gene status. Balt. J. Health Phys. Act. 2019, 11, 26–35. [Google Scholar] [CrossRef]
- Tomczyk, M.; Kortas, J.; Flis, D.; Kaczorowska-Hac, B.; Grzybkowska, A.; Borkowska, A.; Lewicka, E.; Dabrowska-Kugacka, A.; Antosiewicz, J. Marathon Run-induced Changes in the Erythropoietin-Erythroferrone-Hepcidin Axis are Iron Dependent. Int. J. Environ. Res. Public Health 2020, 17, 2781. [Google Scholar] [CrossRef] [Green Version]
- Kortas, J.; Ziemann, E.; Antosiewicz, J. Effect of HFE Gene Mutation on Changes in Iron Metabolism Induced by Nordic Walking in Elderly Women. Clin. Interv. Aging 2020, 15, 663–671. [Google Scholar] [CrossRef]
- Semenova, E.A.; Miyamoto-Mikami, E.; Akimov, E.B.; Al-Khelaifi, F.; Murakami, H.; Zempo, H.; Kostryukova, E.S.; Kulemin, N.A.; Larin, A.K.; Borisov, O.V.; et al. The association of HFE gene H63D polymorphism with endurance athlete status and aerobic capacity: Novel findings and a meta-analysis. Eur. J. Appl. Physiol. 2020, 120, 665–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakkar, D.; Sicova, M.; Guest, N.S.; Garcia-Bailo, B.; El-Sohemy, A. HFE Genotype and Endurance Performance in Competitive Male Athletes. Med. Sci. Sports Exerc. 2021, 53, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Kalpouzos, G.; Mangialasche, F.; Falahati, F.; Laukka, E.J.; Papenberg, G. Contributions of HFE polymorphisms to brain and blood iron load, and their links to cognitive and motor function in healthy adults. Neuropsychopharmacol. Rep. 2021, 41, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Guest, N.S.; Horne, J.; Vanderhout, S.M.; El-Sohemy, A. Sport Nutrigenomics: Personalized Nutrition for Athletic Performance. Front. Nutr. 2019, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.; Farrag, K.; Stein, J. Limitations of Serum Ferritin in Diagnosing Iron Deficiency in Inflammatory Conditions. Int. J. Chronic Dis. 2018, 2018, 9394060. [Google Scholar] [CrossRef] [Green Version]
- Skarpanska-Stejnborn, A.; Basta, P.; Trzeciak, J.; Szczesniak-Pilaczynska, L. Effect of intense physical exercise on hepcidin levels and selected parameters of iron metabolism in rowing athletes. Eur. J. Appl. Physiol. 2015, 115, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, Y.O.; Schmid, A.; Konig, D.; Berg, A. Effects of exercise on soluble transferrin receptor and other variables of the iron status. Br. J. Sports Med. 2002, 36, 195–199. [Google Scholar] [CrossRef] [Green Version]
- Maughan, R.J.; Leiper, J.B.; Bartagi, Z.; Zrifi, R.; Zerguini, Y.; Dvorak, J. Effect of Ramadan fasting on some biochemical and haematological parameters in Tunisian youth soccer players undertaking their usual training and competition schedule. J. Sports Sci. 2008, 26 (Suppl. 3), S39–S46. [Google Scholar] [CrossRef]
- Greenham, G.; Buckley, J.D.; Garrett, J.; Eston, R.; Norton, K. Biomarkers of Physiological Responses to Periods of Intensified, Non-Resistance-Based Exercise Training in Well-Trained Male Athletes: A Systematic Review and Meta-Analysis. Sports Med. 2018, 48, 2517–2548. [Google Scholar] [CrossRef]
- Suominen, P.; Punnonen, K.; Rajamaki, A.; Irjala, K. Serum transferrin receptor and transferrin receptor-ferritin index identify healthy subjects with subclinical iron deficits. Blood 1998, 92, 2934–2939. [Google Scholar] [CrossRef]
- Zügel, M.; Treff, G.; Steinacker, J.M.; Mayer, B.; Winkert, K.; Schumann, U. Increased Hepcidin Levels During a Period of High Training Load Do Not Alter Iron Status in Male Elite Junior Rowers. Front. Physiol. 2020, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.P.R.; Oliveira, R.A.; Silva, E.D.; Lima, G.H.O.; Benetti, M.P.; Kiss, M.A.P.; Sierra, C.A.; Ghorayeb, N.; Seto, J.T.; Pesquero, J.B.; et al. Association Between Hematological Parameters and Iron Metabolism Response After Marathon Race and ACTN3 Genotype. Front. Physiol. 2019, 10, 697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Tok, A.I.Y.; Alagappan, P.; Liedberg, B. Point of care testing of sports biomarkers: Potential applications, recent advances and future outlook. TrAC Trends Anal. Chem. 2021, 142, 116327. [Google Scholar] [CrossRef]
- Peeling, P.; Dawson, B.; Goodman, C.; Landers, G.; Wiegerinck, E.T.; Swinkels, D.W.; Trinder, D. Training surface and intensity: Inflammation, hemolysis, and hepcidin expression. Med. Sci. Sports Exerc. 2009, 41, 1138–1145. [Google Scholar] [CrossRef]
- Peeling, P.; Dawson, B.; Goodman, C.; Landers, G.; Wiegerinck, E.T.; Swinkels, D.W.; Trinder, D. Effects of exercise on hepcidin response and iron metabolism during recovery. Int. J. Sport Nutr. Exerc. Metab. 2009, 19, 583–597. [Google Scholar] [CrossRef] [Green Version]
- Roecker, L.; Meier-Buttermilch, R.; Brechtel, L.; Nemeth, E.; Ganz, T. Iron-regulatory protein hepcidin is increased in female athletes after a marathon. Eur. J. Appl. Physiol. 2005, 95, 569–571. [Google Scholar] [CrossRef]
- Galesloot, T.E.; Vermeulen, S.H.; Geurts-Moespot, A.J.; Klaver, S.M.; Kroot, J.J.; van Tienoven, D.; Wetzels, J.F.; Kiemeney, L.A.; Sweep, F.C.; den Heijer, M.; et al. Serum hepcidin: Reference ranges and biochemical correlates in the general population. Blood 2011, 117, e218–e225. [Google Scholar] [CrossRef]
- Banzet, S.; Sanchez, H.; Chapot, R.; Bigard, X.; Vaulont, S.; Koulmann, N. Interleukin-6 contributes to hepcidin mRNA increase in response to exercise. Cytokine 2012, 58, 158–161. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Chang, Y.Z.; Zhao, B.; Wang, H.T.; Duan, X.L. Does hepatic hepcidin play an important role in exercise-associated anemia in rats? Int. J. Sport Nutr. Exerc. Metab. 2011, 21, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Newlin, M.K.; Williams, S.; McNamara, T.; Tjalsma, H.; Swinkels, D.W.; Haymes, E.M. The Effects of Acute Exercise Bouts on Hepcidin in Women. Int. J. Sport Nutr. Exerc. Metab. 2012, 22, 79. [Google Scholar] [CrossRef]
- Urrutia, P.; Aguirre, P.; Esparza, A.; Tapia, V.; Mena, N.P.; Arredondo, M.; Gonzalez-Billault, C.; Nunez, M.T. Inflammation alters the expression of DMT1, FPN1 and hepcidin, and it causes iron accumulation in central nervous system cells. J. Neurochem. 2013, 126, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chang, Y.; Qian, Z.; Liu, J.; Duan, X. Effects of Swim Training on the Expression of Gastrocnemius’ Iron Transport Proteins in Rats. Chin. J. Sports Med. 2005, 24, 147–151. [Google Scholar] [CrossRef]
- Ingrassia, R.; Garavaglia, B.; Memo, M. DMT1 Expression and Iron Levels at the Crossroads Between Aging and Neurodegeneration. Front. Neurosci. 2019, 13, 575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, D.H.; Kwon, K.C.; Hwang, D.J.; Koo, J.H.; Um, H.S.; Song, H.S.; Kim, J.S.; Jang, Y.; Cho, J.Y. Treadmill Exercise Alleviates Brain Iron Dyshomeostasis Accelerating Neuronal Amyloid-beta Production, Neuronal Cell Death, and Cognitive Impairment in Transgenic Mice Model of Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 3208–3223. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Duan, X.L.; Chang, Y.Z.; Wang, H.T.; Qian, Z.M. Molecular analysis of increased iron status in moderately exercised rats. Mol. Cell. Biochem. 2006, 282, 117–123. [Google Scholar] [CrossRef]
- Wuyun, G.; Hu, Y.; He, Z.; Li, Y.; Yan, X. The Short Tandem Repeat of the DMT1 Gene as a Molecular Marker of Elite Long-Distance Runners. Int. J. Genom. 2019, 2019, 7064703. [Google Scholar] [CrossRef]
- Iolascon, A.; d’Apolito, M.; Servedio, V.; Cimmino, F.; Piga, A.; Camaschella, C. Microcytic anemia and hepatic iron overload in a child with compound heterozygous mutations in DMT1 (SCL11A2). Blood 2006, 107, 349–354. [Google Scholar] [CrossRef] [Green Version]
- Mayr, R.; Janecke, A.R.; Schranz, M.; Griffiths, W.J.; Vogel, W.; Pietrangelo, A.; Zoller, H. Ferroportin disease: A systematic meta-analysis of clinical and molecular findings. J. Hepatol. 2010, 53, 941–949. [Google Scholar] [CrossRef] [Green Version]
- Ka, C.; Guellec, J.; Pepermans, X.; Kannengiesser, C.; Ged, C.; Wuyts, W.; Cassiman, D.; de Ledinghen, V.; Varet, B.; de Kerguenec, C.; et al. The SLC40A1 R178Q mutation is a recurrent cause of hemochromatosis and is associated with a novel pathogenic mechanism. Haematologica 2018, 103, 1796–1805. [Google Scholar] [CrossRef] [Green Version]
- Viveiros, A.; Panzer, M.; Baumgartner, N.; Schaefer, B.; Finkenstedt, A.; Henninger, B.; Theurl, I.; Nachbaur, K.; Weiss, G.; Haubner, R.; et al. Reduced iron export associated with hepcidin resistance can explain the iron overload spectrum in ferroportin disease. Liver Int. 2020, 40, 1941–1951. [Google Scholar] [CrossRef]
- Xie, J.; Wang, Y.; Freeman, M.E., III; Barlogie, B.; Yi, Q. β2-Microglobulin as a negative regulator of the immune system: High concentrations of the protein inhibit in vitro generation of functional dendritic cells. Blood 2003, 101, 4005–4012. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, E.B.; Mogensen, C.E.; Larsen, J.S. Effects of exercise on urinary excretion of albumin and β2-microglobulin in young patients with mild essential hypertension without treatment and during long-term propranolol treatment. Scand. J. Clin. Lab. Investig. 1981, 41, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, D.A.; Méndez, C.H.; Angulo, O.J.; Cruz, J.A.; Chaves, J.L.; Vargas-Molina, S.; Petro, J.L.; Palma, L.H. Efectos del ejercicio grupal de moderada intensidad sobre la presión arterial y capacidad funcional en mujeres posmenopáusicas hipertensas. RICYDE. Rev. Int. Cienc. Del Deporte 2021, 17, 306–320. [Google Scholar] [CrossRef]
- Li, L.; Dong, M.; Wang, X.G. The Implication and Significance of Beta 2 Microglobulin: A Conservative Multifunctional Regulator. Chin. Med. J. 2016, 129, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, X.; Zu, Y.; Li, H.; Wang, S. Prognostic value of beta-2 microglobulin on mortality in chronic kidney disease patients: A systematic review and meta-analysis. Ther. Apher. Dial. 2021, 26, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.M.; Kimura, E.; Harada, R.K.; Nair, N.; Narasimhan, B.; Meng, X.Y.; Zhang, F.; Beck, K.R.; Olin, J.W.; Fung, E.T.; et al. Beta2-microglobulin as a biomarker in peripheral arterial disease: Proteomic profiling and clinical studies. Circulation 2007, 116, 1396–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Liu, B.; Wei, J. Beta2-microglobulin(B2M) in cancer immunotherapies: Biological function, resistance and remedy. Cancer Lett. 2021, 517, 96–104. [Google Scholar] [CrossRef]
- Althubiti, M.; Elzubier, M.; Alotaibi, G.S.; Althubaiti, M.A.; Alsadi, H.H.; Alhazmi, Z.A.; Alghamdi, F.; El-Readi, M.Z.; Almaimani, R.; Babakr, A. Beta 2 microglobulin correlates with oxidative stress in elderly. Exp. Gerontol. 2021, 150, 111359. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, C.P.; Chen, S.S.; Ng, Y.H.; Roumelioti, M.E.; Shaffi, K.; Singh, P.P.; Tzamaloukas, A.H. Rediscovering Beta-2 Microglobulin As a Biomarker across the Spectrum of Kidney Diseases. Front. Med. 2017, 4, 73. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.V.; Chen, Y.C.; Wu, W.T.; Shen, H.J.; Huang, K.C.; Chu, H.P.; Han, D.S. Expression of Telomeric Repeat-Containing RNA Decreases in Sarcopenia and Increases after Exercise and Nutrition Intervention. Nutrients 2020, 12, 3766. [Google Scholar] [CrossRef]
- Mahoney, D.J.; Carey, K.; Fu, M.H.; Snow, R.; Cameron-Smith, D.; Parise, G.; Tarnopolsky, M.A. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol. Genom. 2004, 18, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Nihon-Yanagi, Y.; Terai, K.; Murano, T.; Kawai, T.; Kimura, S.; Okazumi, S. beta-2 microglobulin is unsuitable as an internal reference gene for the analysis of gene expression in human colorectal cancer. Biomed. Rep. 2013, 1, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Umemoto, T.; Tanaka, Y.; Okano, T.; Yamato, M. beta2-Microglobulin is an appropriate reference gene for RT-PCR-based gene expression analysis of hematopoietic stem cells. Regen 2015, 1, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Collins, J.F. Copper. In Present Knowledge in Nutrition; Marriott, B.P., Birt, D.F., Stallings, V.A., Yates, A.A., Eds.; Academic Press: New York, NY, USA, 2020; Volume 1. [Google Scholar]
- Sheikh, N.; Dudas, J.; Ramadori, G. Changes of gene expression of iron regulatory proteins during turpentine oil-induced acute-phase response in the rat. Lab. Investig. 2007, 87, 713–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, T.; Ali, G.; Sheikh, N. Immunosuppressant-Induced Oxidative Stress and Iron: A Paradigm Shift from Systemic to Intrahepatic Abnormalities. Oxid. Med. Cell. Longev. 2020, 2020, 8675275. [Google Scholar] [CrossRef]
- Zacchi, P.; Belmonte, B.; Mangogna, A.; Morello, G.; Scola, L.; Martorana, A.; Borelli, V. The Ferroxidase Hephaestin in Lung Cancer: Pathological Significance and Prognostic Value. Front. Oncol. 2021, 11, 638856. [Google Scholar] [CrossRef]
- Forero, D.A.; Curioso, W.H.; Patrinos, G.P. The importance of adherence to international standards for depositing open data in public repositories. BMC Res. Notes 2021, 14, 405. [Google Scholar] [CrossRef]
- DeRosa, A.; Leftin, A. The Iron Curtain: Macrophages at the Interface of Systemic and Microenvironmental Iron Metabolism and Immune Response in Cancer. Front. Immunol. 2021, 12, 614294. [Google Scholar] [CrossRef]
- Mertens, C.; Marques, O.; Horvat, N.K.; Simonetti, M.; Muckenthaler, M.U.; Jung, M. The Macrophage Iron Signature in Health and Disease. Int. J. Mol. Sci. 2021, 22, 8457. [Google Scholar] [CrossRef]
- Gomes, A.C.; Moreira, A.C.; Mesquita, G.; Gomes, M.S. Modulation of Iron Metabolism in Response to Infection: Twists for All Tastes. Pharmaceuticals 2018, 11, 84. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Du, Y.; Wang, J. Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provides potential targets for mechanism research. Sci. Rep. 2015, 5, 12939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suman, S.; Mishra, A. An interaction network driven approach for identifying biomarkers for progressing cervical intraepithelial neoplasia. Sci. Rep. 2018, 8, 12927. [Google Scholar] [CrossRef]
- Oany, A.R.; Mia, M.; Pervin, T.; Alyami, S.A.; Moni, M.A. Integrative Systems Biology Approaches to Identify Potential Biomarkers and Pathways of Cervical Cancer. J. Pers. Med. 2021, 11, 363. [Google Scholar] [CrossRef]
- Kong, W.N.; Niu, Q.M.; Ge, L.; Zhang, N.; Yan, S.F.; Chen, W.B.; Chang, Y.Z.; Zhao, S.E. Sex differences in iron status and hepcidin expression in rats. Biol. Trace Elem. Res. 2014, 160, 258–267. [Google Scholar] [CrossRef]
- Pedlar, C.R.; Brugnara, C.; Bruinvels, G.; Burden, R. Iron balance and iron supplementation for the female athlete: A practical approach. Eur. J. Sport Sci. 2018, 18, 295–305. [Google Scholar] [CrossRef]
- Badenhorst, C.E.; Goto, K.; O’Brien, W.J.; Sims, S. Iron status in athletic females, a shift in perspective on an old paradigm. J. Sports Sci. 2021, 39, 1565–1575. [Google Scholar] [CrossRef]
- Magliulo, L.; Bondi, D.; Pietrangelo, T.; Fulle, S.; Piccinelli, R.; Jandova, T.; Blasio, G.D.; Taraborrelli, M.; Verratti, V. Serum ferritin and vitamin D evaluation in response to high altitude comparing Italians trekkers vs Nepalese porters. Eur. J. Sport Sci. 2021, 21, 994–1002. [Google Scholar] [CrossRef]
- Hu, W.J.; Zhou, S.M.; Yang, J.S.; Meng, F.G. Computational simulations to predict creatine kinase-associated factors: Protein-protein interaction studies of brain and muscle types of creatine kinases. Enzym. Res. 2011, 2011, 328249. [Google Scholar] [CrossRef] [Green Version]
- Zvyagin, I.V.; Tsvetkov, V.O.; Chudakov, D.M.; Shugay, M. An overview of immunoinformatics approaches and databases linking T cell receptor repertoires to their antigen specificity. Immunogenetics 2020, 72, 77–84. [Google Scholar] [CrossRef]






| Recommended Name (Alternative Names) | Gene Name (Location) | Ensembl ID | Protein Features (UniProtKB/PDB Entry) | Cellular Location | Molecular Function | Protein Expression * (BioGPS ID) |
|---|---|---|---|---|---|---|
| Cytochrome b reductase 1 (Duodenal cytochrome b; Ferric-chelate reductase 3) | CYBRD1 (2q31.1) | ENSG00000071967 | Length: 286 Mass: 31,641 Da (Q53TN4/5ZLE) | Integral component of membrane. Present at the brush border of duodenal enterocytes where it probably reduces dietary Fe3+ thereby facilitating its transport into the mucosal cells. | Ferric-chelate reductase that reduces Fe3+ to Fe2+. Uses ascorbate as electron donor. May be involved in extracellular ascorbate recycling in erythrocyte membranes. May also act as a ferrireductase in airway epithelial cells. | Thyroid gland, small intestine, colon, testis, gallbladder, ovary, breast endometrium (79901) |
| Natural resistance- associated macrophage protein 2—NRAM2 (Solute carrier family 11 member 2; Divalent metal ion transporter 1 [DMT1]) | SLC11A2 (12q13.12) | ENSG00000110911 | Length: 568 Mass: 62,266 Da (P49281/5F0L) | Integral component of plasma membrane. Present at the apical plasma membrane where it is involved in Fe uptake into duodenal enterocytes. May serve to import Fe into the mitochondria. | Important in metal transport, in particular Fe. Can also transport manganese, cobalt, cadmium, nickel, vanadium and lead. May play an important role in hepatic Fe accumulation and tissue Fe distribution. | Salivary gland, cerebral cortex, adrenal gland, bronchus, lung, stomach, colon, rectum, liver, gallbladder, pancreas, kidney (4891) |
| Proton-coupled folate transporter (Heme carrier protein 1) | SLC46A1 (17q11.2) | ENSG00000076351 | Length: 459 Mass: 49,771 Da (Q96NT5/-) | Apical plasma membrane. Localizes to the apical membrane of intestinal cells in Fe-deficient cells, while it resides in the cytoplasm in Fe-replete cells. | It has been shown to act both as an intestinal proton-coupled high-affinity folate transporter and as an intestinal heme transporter, which mediates heme uptake from the gut lumen into duodenal epithelial cells. | Testis, small intestine, duodenum, colon (113235) |
| Scavenger receptor cysteine-rich type 1 protein † (Hemoglobin scavenger receptor) | CD163 (12p13.31) | ENSG00000177575 | Length: 1156 Mass: 125,45 Da (Q86VB7/-SWISS-MODEL Repository Q86VB7) | Extracellular region or secreted and plasma membrane. Acute phase-regulated receptor involved in clearance and endocytosis of hemoglobin/haptoglobin complexes. | May play a role in the uptake and recycling of Fe, via endocytosis of hemoglobin/haptoglobin and subsequent breakdown of heme. Binds hemoglobin/haptoglobin complexes in a calcium-dependent and pH-dependent manner. | Lung, spleen, bone marrow, lymph node, appendix, tonsil (9332) |
| Heme oxygenase 1 —HMOX-1 | HMOX1 (22q12.3) | ENSG00000100292 | Length: 288 Mass: 32,219 Da (P09601/1N3U) | Endoplasmic reticulum membrane and perinuclear region of cytoplasm. Under physiological conditions, the activity of heme oxygenase is highest in the spleen, where senescent erythrocytes are sequestrated and destroyed. | Heme oxygenase cleaves the heme ring at the alpha methene bridge to form biliverdin. Biliverdin is subsequently converted to bilirubin by biliverdin reductase. | Lung, duodenum, small intestine, spleen, bone marrow, placenta, appendix, lymph node, tonsil (3162) |
| Ferritin heavy chain | FTH1 (11q12.3) | ENSG00000167996 | Length: 183 Mass: 21,226 Da (P02794/1FHA) | Cytosol, extracellular exosome, autolysosome, protoplasm. | Stores Fe in a soluble, non-toxic, readily available form. Has ferroxidase activity. Fe is taken up in the ferrous form and deposited as ferric hydroxides after oxidation. Also plays a role in delivery of Fe to cells. | Cerebral cortex, bone marrow, hippocampus, small intestine (2495) |
| Ferritin light chain | FTL (19q13.33) | ENSG00000087086 | Length: 175 Mass: 20,020 Da (P02792/2FFX) | Cerebral cortex, cerebellum, lung, liver, kidney (2512) | ||
| Scavenger receptor class A member 5— (Ferritin receptor) † | SCARA5 (8p21.1) | ENSG00000168079 | Length: 495 Mass: 53,994 Da (Q6ZMJ2/- SWISS-MODEL Repository Q6ZMJ2) | Integral component of plasma membrane. | Ferritin receptor that mediates non-transferrin-dependent delivery of Fe. Mediates cellular uptake of ferritin-bound Fe by stimulating ferritin endocytosis from the cell surface with consequent Fe delivery within the cell. | Adrenal gland, stomach, small intestine, colon, rectum, tonsil, gallbladder, lymph node, (286133) |
| Poly (rC)–binding proteins | PCBP2 (12q13.13) | ENSG00000197111 | Length: 365 Mass: 38,580 Da (Q15366/2AXY) | Cytosol, extracellular region or secreted, nucleus and other cell locations. | As a chaperone, promotes intracellular Fe flux. It can directly receive Fe2+ from CYBRD1 or transfer Fe to the Fe2+ exporter, SLC40A1. | Cerebellum, bronchus, oral mucosa, stomach, liver, testis, kidney (5094) |
| Solute carrier family 40 member 1 (Ferroportin 1) | SLC40A1 (2q32.2) | ENSG00000138449 | Length: 571 Mass: 62,542 Da (Q9NP59/-) | Basolateral plasma membrane and integral component of plasma membrane. | May be involved in Fe export from duodenal epithelial cell and in transfer of Fe between maternal and fetal circulation. Mediates Fe efflux in the presence of a ferroxidase (hephaestin and/or ceruloplasmin). | Bone marrow, duodenum, small intestine, smooth muscle, skeletal muscle (30061) |
| Hephaestin | HEPH (Xq12) | ENSG00000089472 | Length: 1158 Mass: 130,44 Da (Q9BQS7/- SWISS-MODEL Repository Q9BQS7) | Basolateral plasma membrane and integral component of plasma membrane. | May function as a ferroxidase for Fe2+ to Fe3+ conversion and may be involved in copper transport and metabolism. Implicated in [Fe] regulation and may mediate Fe efflux associated with SLC40A1. | Cerebral cortex, hippocampus, thyroid gland, lungs, stomach, small intestine, liver, pancreas, (9843) |
| Ceruloplasmin | CP (3q24-q25.1) | ENSG00000047457 | Length: 1065 Mass: 122,20 Da (P00450/1KCW) | Extracellular region or secreted, plasma membrane, endoplasmic reticulum lumen and lysosomal membrane. | Ceruloplasmin is a blue, copper-binding (6–7 atoms per molecule) glycoprotein. It has ferroxidase activity oxidizing Fe2+ to Fe3+ without releasing radical oxygen species. It is involved in Fe transport across the cell membrane. | The RNA is highly expressed in the liver. Mainly found in the bloodstream (1356) |
| Transferrin | TF (3q22.1) | ENSG00000091513 | Length: 698 Mass: 77,064 Da (P02787/1A8E) | Secreted. Blood microparticle, extracellular exosome, and extrinsic component of external side of plasma membrane. | Transferrins are Fe-binding transport proteins that can bind two Fe3+ ions in association with the binding of an anion, usually bicarbonate. They are responsible for the transport of Fe from sites of absorption and heme degradation to those of storage and utilization. | The RNA is highly expressed in the liver. Protein is highly expressed in placenta and testis, although is mainly found in the bloodstream (7018) |
| Transferrin receptor protein 1 | TFRC (3q29) | ENSG00000072274 | Length: 760 Mass: 84,871 Da (P02786/1CX8) | Integral component of plasma membrane, endosome (clathrin-coated vesicle membrane), blood microparticle. Positively regulates T and B cell proliferation through Fe uptake. | Cellular uptake of Fe occurs via receptor-mediated endocytosis of ligand-occupied transferrin receptor into specialized endosomes. Endosomal acidification leads to Fe release. The apotransferrin-receptor complex is then recycled to the cell surface with a return to neutral pH and the concomitant loss of affinity of apotransferrin for its receptor. | Placenta, bone marrow, cerebellum, hippocampus, adrenal gland, bronchus, lung, oral mucosa, esophagus, duodenum, colon, rectum, urinary bladder, testis (7037) |
| Hepcidin | HAMP (19q13.12) | ENSG00000105697 | Length: 84 Mass: 9408 Da (P81172/1M4E) | Extracellular region or secreted. Controls the major flows of Fe into plasma: absorption of dietary Fe in the intestine, recycling of Fe by macrophages, which phagocytose old erythrocytes and other cells, and mobilization of stored Fe from hepatocytes. | Liver-produced hormone that constitutes the main circulating regulator of Fe absorption and distribution across tissues. Acts by promoting endocytosis and degradation of SLC40A1 (ferroportin), leading to the retention of Fe in Fe-exporting cells and decreased flow of Fe into plasma. | The RNA is highly expressed in the liver although very low expression levels are found in heart muscle and spinal cord. Mainly found in the bloodstream. (57817) |
| Iron-responsive element-binding protein 2 † (Iron regulatory protein 2) | IREB2 (15q25.1) | ENSG00000136381 | Length: 963 Mass: 105,05 Da (P48200/ SWISS-MODEL Repository P48200) | Cytoplasm, mitochondrion. | RNA-binding protein that binds to iron-responsive elements (IRES), which are stem-loop structures found in the 5′-UTR of ferritin, and delta aminolevulinic acid synthase mRNAs, and in the 3′-UTR of transferrin receptor mRNA. | Cerebellum, parathyroid gland, adrenal gland, oral mucosa, stomach, small intestine, kidney, prostate (3658) |
| Biological Process (GO) | ||
|---|---|---|
| GO-term | Description | FDR p-value |
| GO:0055072 | iron ion homeostasis | 9.58 × 10−31 |
| GO:0006879 | cellular iron ion homeostasis | 5.91 × 10−30 |
| GO:0006826 | iron ion transport | 3.30 × 10−20 |
| GO:0000041 | transition metal ion transport | 7.82 × 10−20 |
| GO:0019725 | cellular homeostasis | 6.48 × 10−16 |
| Molecular Function (GO) | ||
| GO-term | Description | FDR p-value |
| GO:0005381 | iron ion transmembrane transporter activity | 5.37 × 10−7 |
| GO:0016722 | oxidoreductase activity, oxidizing metal ions | 7.59 × 10−7 |
| GO:0042605 | peptide antigen binding | 1.94 × 10−6 |
| GO:0004322 | ferroxidase activity | 7.06 × 10−6 |
| GO:0046977 | TAP binding | 7.06 × 10−6 |
| Cellular Component (GO) | ||
| GO-term | Description | FDR p-value |
| GO:0055037 | recycling endosome | 1.41 × 10−10 |
| GO:0042612 | MHC class I protein complex | 3.39 × 10−10 |
| GO:0005769 | early endosome | 1.94 × 10−8 |
| GO:0009986 | cell surface | 2.09 × 10−8 |
| GO:1990712 | HFE-transferrin receptor complex | 3.87 × 10−8 |
| KEGG Pathways | ||
| Pathway ID | Description | FDR p-value |
| hsa04978 | mineral absorption | 1.25 × 10−15 |
| hsa04216 | ferroptosis | 1.75 × 10−14 |
| hsa04612 | antigen processing and presentation | 3.69 × 10−7 |
| hsa05330 | allograft rejection | 1.60 × 10−6 |
| hsa05332 | graft-versus-host disease | 1.60 × 10−6 |
| PFAM Protein Domains | ||
| Domain | Description | FDR p-value |
| PF07654 | immunoglobulin C1-set domain | 1.98 × 10−9 |
| PF06623 | MHC_I C-terminus | 2.78 × 10−9 |
| PF00129 | Class I histocompatibility antigen, domains alpha 1 and 2 | 2.78 × 10−9 |
| PF00210 | ferritin-like domain | 0.00025 |
| PF07731 | multicopper oxidase | 0.00025 |
| Protein Name | Degree Centrality | Betweenness Centrality | Eigenvector Centrality | Subgraph Centrality | Average Score † |
|---|---|---|---|---|---|
| HFE | 22 | 44.85 | 1.00 | 215,209.80 | 0.831272727 |
| TFRC | 20 | 30.41 | 0.99 | 195,055.82 | 0.9057 |
| B2M | 18 | 38.04 | 0.78 | 97,724.47 | 0.955555556 |
| SLC11A2 | 18 | 16.37 | 0.88 | 178,173.90 | 0.852666667 |
| FTH1 | 16 | 34.26 | 0.64 | 87,931.41 | 0.872375 |
| HEPH | 16 | 35.32 | 0.78 | 140,287.68 | 0.857 |
| SLC40A1 | 16 | 7.12 | 0.85 | 159,359.04 | 0.882625 |
| CP | 12 | 15.58 | 0.52 | 54,947.33 | 0.906166667 |
| HAMP | 12 | 0.33 | 0.73 | 111,523.40 | 0.889166667 |
| CYBRD1 | 10 | 0.00 | 0.55 | 78,214.76 | 0.8214 |
| Protein | ImmunomeBase IKB | InnateDB Interactions | BiomarkerBase™ | MarkerDB | Normal | Abnormal | Exercise | |
|---|---|---|---|---|---|---|---|---|
| CTs | Conditions | |||||||
| HFE | Yes | 16 | 19 | 988 | Yes * | G/G C/C | C282Y (A/G, A/A) H63D (C/G, G/G) | ★★★★★ |
| TFRC | Yes | 73 | 146 | 849 | Yes | F †: 1.9–4.4 mg·L−1 M †: 2.2–5 mg·L−1 | F: >4.4 mg·L−1 M: >5 mg·L−1 | ★★★★★ |
| B2M | Yes | 188 | 176 | 946 | Yes | 1.21–2.7 μg·mL−1 | >4 μg·mL−1 | ★★ |
| SLC11A2 | No | 8 | 3 | 338 | No | 258/258 bp and 258 bp alleles overrepresented in athletes | ★★★ | |
| FTH1 | Yes | 54 | 886 | 1176 | Yes | F: 11–307 μg·L−1 M: 24–336 μg·L−1 | F: <11 μg·L−1 M: <24 μg·L−1 | ★★★★★ |
| HEPH | No | - | 0 | 115 | No | NA | NA | ? |
| SLC40A1 | No | - | 9 | 513 | Yes * | C/C | R178Q (C/T) | ★★★ |
| CP | No | 7 | 53 | 1020 | Yes | 200–350 mg·L−1 | <200 mg·L−1 | ? |
| HAMP | Yes | 5 | 173 | 669 | Yes * | C/C F: 1–4.1 nM F ‡: 3.2–8.5 nM M: 1–7.8 nM | C72Ter (C/A, C/T) >8.5 nM | ★★★★★ |
| CYBRD1 | No | 3 | 0 | 59 | No | NA | NA | ? |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonilla, D.A.; Moreno, Y.; Petro, J.L.; Forero, D.A.; Vargas-Molina, S.; Odriozola-Martínez, A.; Orozco, C.A.; Stout, J.R.; Rawson, E.S.; Kreider, R.B. A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression. Biomedicines 2022, 10, 724. https://doi.org/10.3390/biomedicines10030724
Bonilla DA, Moreno Y, Petro JL, Forero DA, Vargas-Molina S, Odriozola-Martínez A, Orozco CA, Stout JR, Rawson ES, Kreider RB. A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression. Biomedicines. 2022; 10(3):724. https://doi.org/10.3390/biomedicines10030724
Chicago/Turabian StyleBonilla, Diego A., Yurany Moreno, Jorge L. Petro, Diego A. Forero, Salvador Vargas-Molina, Adrián Odriozola-Martínez, Carlos A. Orozco, Jeffrey R. Stout, Eric S. Rawson, and Richard B. Kreider. 2022. "A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression" Biomedicines 10, no. 3: 724. https://doi.org/10.3390/biomedicines10030724
APA StyleBonilla, D. A., Moreno, Y., Petro, J. L., Forero, D. A., Vargas-Molina, S., Odriozola-Martínez, A., Orozco, C. A., Stout, J. R., Rawson, E. S., & Kreider, R. B. (2022). A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression. Biomedicines, 10(3), 724. https://doi.org/10.3390/biomedicines10030724