Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. Stable Transfection
2.3. Cell Morphology Analysis
2.4. Transwell Migration and Invasion Assay
2.5. MMP-7 ELISA Assay
2.6. Protein Extraction and Western Blotting
2.7. siRNA Transfection
2.8. Statistical Analysis
3. Results
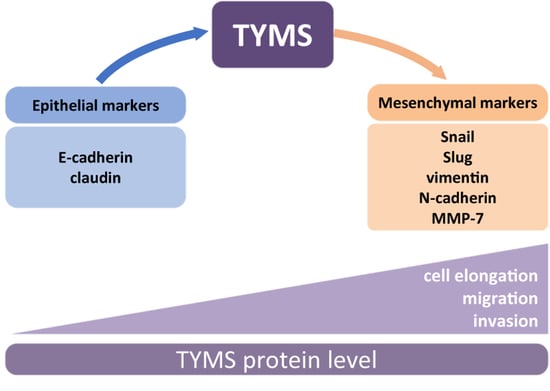
3.1. TYMS Protein Level Corresponds with Invasive Potency of Colon Cancer Cells
3.2. TYMS Overexpression Induce Epithelial–Mesenchymal Transition in Colon Cancer Cells
3.3. TYMS Overexpression Induce Invasiveness in Colon Cancer Cells
3.4. TYMS Downregulation Abolishes the EMT Process and Inhibits Colon Cancer Cells Invasion
3.5. TYMS Silencing Reverses Invasive Phenotype in TYMS-Overexpressing Cancer
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 2020, 206, 107447. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.J.; Backus, H.H.; Freemantle, S.; van Triest, B.; Codacci-Pisanelli, G.; van der Wilt, C.L.; Smid, K.; Lunec, J.; Calvert, A.H.; Marsh, S.; et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim. Biophys. Acta 2002, 1587, 194–205. [Google Scholar] [CrossRef] [Green Version]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Jiao, Y.; Li, Y.; Ji, B.; Jia, B.; Liu, B. Tyms presents a novel biomarker for diagnosis and prognosis in patients with pancreatic cancer. Medicine 2019, 98, e18487. [Google Scholar] [CrossRef]
- Nomura, T.; Nakagawa, M.; Fujita, Y.; Hanada, T.; Mimata, H.; Nomura, Y. Clinical significance of thymidylate synthase expression in bladder cancer. Int. J. Urol. 2002, 9, 368–376. [Google Scholar] [CrossRef]
- Song, S.; Tian, B.; Zhang, M.; Gao, X.; Jie, L.; Liu, P.; Li, J. Diagnostic and prognostic value of thymidylate synthase expression in breast cancer. Clin. Exp. Pharmacol. Physiol. 2021, 48, 279–287. [Google Scholar] [CrossRef]
- Lu, Y.; Zhuo, C.; Cui, B.; Liu, Z.; Zhou, P.; Lu, Y.; Wang, B. Tyms serves as a prognostic indicator to predict the lymph node metastasis in Chinese patients with colorectal cancer. Clin. Biochem. 2013, 46, 1478–1483. [Google Scholar] [CrossRef]
- Kimura, M.; Kuwabara, Y.; Mitsui, A.; Ishiguro, H.; Sugito, N.; Tanaka, T.; Shiozaki, M.; Naganawa, Y.; Takeyama, H. Thymidylate synthetase and dihydropyrimidine dehydrogenase mrna levels in esophageal cancer. Oncol. Lett. 2011, 2, 297–301. [Google Scholar] [CrossRef]
- Siddiqui, A.; Gollavilli, P.N.; Schwab, A.; Vazakidou, M.E.; Ersan, P.G.; Ramakrishnan, M.; Pluim, D.; Coggins, S.; Saatci, O.; Annaratone, L.; et al. Thymidylate synthase maintains the de-differentiated state of triple negative breast cancers. Cell Death Differ. 2019, 26, 2223–2236. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, A.; Vazakidou, M.E.; Schwab, A.; Napoli, F.; Fernandez-Molina, C.; Rapa, I.; Stemmler, M.P.; Volante, M.; Brabletz, T.; Ceppi, P. Thymidylate synthase is functionally associated with zeb1 and contributes to the epithelial-to-mesenchymal transition of cancer cells. J. Pathol. 2017, 242, 221–233. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Gollavilli, P.N.; Ramesh, V.; Parma, B.; Schwab, A.; Vazakidou, M.E.; Natesan, R.; Saatci, O.; Rapa, I.; Bironzo, P.; et al. Thymidylate synthase drives the phenotypes of epithelial-to-mesenchymal transition in non-small cell lung cancer. Br. J. Cancer 2020, 124, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Goodall, G.J. The many regulators of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2022, 23, 89–90. [Google Scholar] [CrossRef] [PubMed]
- Heerboth, S.; Housman, G.; Leary, M.; Longacre, M.; Byler, S.; Lapinska, K.; Willbanks, A.; Sarkar, S. Emt and tumor metastasis. Clin. Transl. Med. 2015, 4, e6. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Muthusamy, B.P.; Saeteurn, K.Y. Signaling pathway cooperation in TGF-β-induced epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2014, 31, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The role of snail in emt and tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef]
- Sobierajska, K.; Wieczorek, K.; Ciszewski, W.M.; Sacewicz-Hofman, I.; Wawro, M.E.; Wiktorska, M.; Boncela, J.; Papiewska-Pajak, I.; Kwasniak, P.; Wyroba, E.; et al. β-III tubulin modulates the behavior of snail overexpressed during the epithelial-to-mesenchymal transition in colon cancer cells. Biochim. Biophys. Acta 2016, 1863, 2221–2233. [Google Scholar] [CrossRef]
- Sobierajska, K.; Ciszewski, W.M.; Wawro, M.E.; Wieczorek-Szukala, K.; Boncela, J.; Papiewska-Pajak, I.; Niewiarowska, J.; Kowalska, M.A. Tubb4b downregulation is critical for increasing migration of metastatic colon cancer cells. Cells 2019, 8, 810. [Google Scholar] [CrossRef] [Green Version]
- Sobierajska, K.; Wawro, M.E.; Ciszewski, W.M.; Niewiarowska, J. Transforming Growth Factor-β Receptor Internalization via Caveolae Is Regulated by Tubulin-β2 and Tubulin-β3 during Endothelial-Mesenchymal Transition. Am. J. Pathol. 2019, 189, 2531–2546. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-mesenchymal transition in cancer: A historical overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Zeng, Z.S.; Shu, W.P.; Cohen, A.M.; Guillem, J.G. Matrix metalloproteinase-7 expression in colorectal cancer liver metastases: Evidence for involvement of mmp-7 activation in human cancer metastases. Clin. Cancer Res. 2002, 8, 144–148. [Google Scholar]
- Said, A.H.; Raufman, J.P.; Xie, G. The role of matrix metalloproteinases in colorectal cancer. Cancers 2014, 6, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Javarsiani, M.H.; Javanmard, S.H.; Colonna, F. Metastatic components in colorectal cancer. J. Res. Med. Sci. 2019, 24, 75. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, C.M.; Krauss, J.C. Emerging systemic therapies for colorectal cancer. Clin. Colon Rectal Surg. 2018, 31, 179–191. [Google Scholar] [PubMed]
- Sethy, C.; Kundu, C.N. 5-fluorouracil (5-fu) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed. Pharm. 2021, 137, 111285. [Google Scholar] [CrossRef] [PubMed]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef]
- Johnston, P.G.; Lenz, H.J.; Leichman, C.G.; Danenberg, K.D.; Allegra, C.J.; Danenberg, P.V.; Leichman, L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995, 55, 1407–1412. [Google Scholar]
- Parr, A.L.; Drake, J.C.; Gress, R.E.; Schwartz, G.; Steinberg, S.M.; Allegra, C.J. 5-fluorouracil-mediated thymidylate synthase induction in malignant and nonmalignant human cells. Biochem. Pharmacol. 1998, 56, 231–235. [Google Scholar] [CrossRef]
- Chu, E.; Koeller, D.M.; Johnston, P.G.; Zinn, S.; Allegra, C.J. Regulation of thymidylate synthase in human colon cancer cells treated with 5-fluorouracil and interferon-gamma. Mol. Pharmacol. 1993, 43, 527–533. [Google Scholar]
- Kamoshida, S.; Matsuoka, H.; Ishikawa, T.; Maeda, K.; Shimomura, R.; Inada, K.; Tsutsumi, Y. Immunohistochemical evaluation of thymidylate synthase (TS) and p16ink4a in advanced colorectal cancer: Implication of TS expression in 5-fu-based adjuvant chemotherapy. Jpn. J. Clin. Oncol. 2004, 34, 594–601. [Google Scholar] [CrossRef]
- Kang, M.; Zheng, W.; Chen, Q.; Qin, W.; Li, P.; Huang, S.; Zhou, Y.; Wang, L.; Cai, H.; Lu, W.; et al. Thymidylate synthase prompts metastatic progression through the dTMP associated EMT process in pancreatic ductal adenocarcinoma. Cancer Lett. 2018, 419, 40–52. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciszewski, W.M.; Chmielewska-Kassassir, M.; Wozniak, L.A.; Sobierajska, K. Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells. Biomedicines 2022, 10, 1267. https://doi.org/10.3390/biomedicines10061267
Ciszewski WM, Chmielewska-Kassassir M, Wozniak LA, Sobierajska K. Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells. Biomedicines. 2022; 10(6):1267. https://doi.org/10.3390/biomedicines10061267
Chicago/Turabian StyleCiszewski, Wojciech M., Małgorzata Chmielewska-Kassassir, Lucyna A. Wozniak, and Katarzyna Sobierajska. 2022. "Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells" Biomedicines 10, no. 6: 1267. https://doi.org/10.3390/biomedicines10061267
APA StyleCiszewski, W. M., Chmielewska-Kassassir, M., Wozniak, L. A., & Sobierajska, K. (2022). Thymidylate Synthase Overexpression Drives the Invasive Phenotype in Colon Cancer Cells. Biomedicines, 10(6), 1267. https://doi.org/10.3390/biomedicines10061267