Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2—New Data for a Possible Role in the Long COVID Pattern
Abstract
:1. Introduction
2. Materials and Methods
- Culturing samples described in Petrillo et al. [34]: samples called A are the cultures of stool bacteria from COVID-19 patients; samples called B(A+) are the cultures of stool bacteria from healthy people but contaminated with the supernatant from samples A; samples called C are the cultures of bacteria collected and grown after centrifuge of samples A and removal of the supernatant. Samples neg-B are the cultures of stool bacteria of healthy people that are the negative control. Moreover, an increase of RNA viral load up to day 30 of cultures in samples A and samples B(A+), and how some antibiotics determine the decrease of viral RNA load in the cultures, was reported; in particular see Table 1. In addition, on aliquots of these cultures, the proteomic exams with the matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) technique and the surface-activated chemical-ionization (SACI) approach [34,35,36,37,38,39] were performed, as just described in [32], searching the unique new molecules that we have previously found in the plasma and urine of COVID-19 patients. The bacteria culture controls, derived from healthy persons, were negative for the increase of RNA viral load as previously described [34] and also for toxin-like peptides presence now reported. All patients gave their consent in accordance with Italian legislation.
- Mass spectrometry data acquisition at different time points (beginning of culturing, after 7, 14, 21, 30 days) by means of Cloud ion mobility mass spectrometry (CIMS) coupled with surface-Electrospray-NIST-activated chemical ionization (SANS), followed by Surface Activated Chemical Ionization—Electrospray—NIST Bayesian model search (SANIST-CIMS) against the complete ‘Uni-Prot KB set of manually revised venom proteins and toxins’ [40] mixed with a subset of non-venom proteins and toxins from UniProt KB to give statistical significance to the results for the presence of proteins with potentially toxic effects.
- Repetition of mass spectrometry data acquisition in the 18 aliquots derived from sample B(A+) at day 21, where antibiotic tests were performed and consisting in the addition of a specific molecule (each of the following: metronidazole, clindamycin, lincomycin, piperacillin+tazobactam, vancomycin, amoxicillin, ampicillin, cefixime, ceftriaxone, meropenem, rifaximin, azithromycin, erythromycin, gentamicin, ciprofloxacin, colistin, levofloxacin, and teicoplanin), for detail see Table 1, previously described in [34].
- Spectral counting [41] was performed in every aliquot, considering the toxin-like peptides abundance respect the culture-negative from SARS-CoV-2 derived from healthy patients. Spectral counting is a semiquantitative mass spectrometry approach for defining the abundance of the molecules under study. The spectral counting parameter was obtained using the exponentially modified protein abundance index (emPAI) [42] approach corrected by a nonparametric normalization index.
- In order to verify the reproducibility of our results, the whole experiment was repeated three times independently.
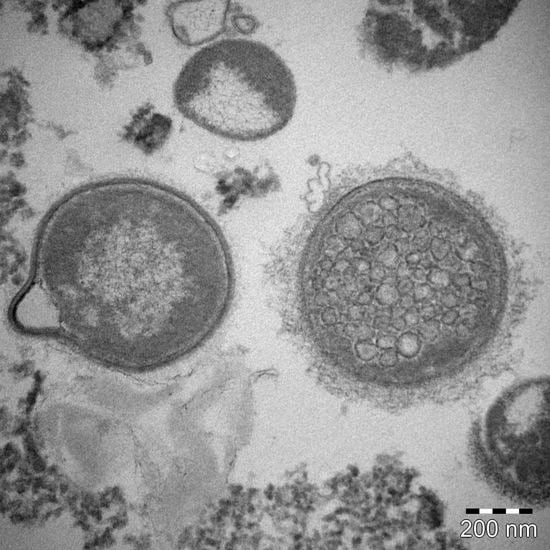
3. New Data Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620, Erratum in Nat. Commun. 2021, 12, 2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Harapan, B.N.; Yoo, H.J. Neurological symptoms, manifestations, and complications associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19). J. Neurol. 2021, 268, 3059–3071. [Google Scholar] [CrossRef] [PubMed]
- Gromova, O.A.; Torshin, I.Y.; Semenov, V.A.; Putilina, M.V.; Chuchalin, A.G. Direct and Indirect Neurological Signs of COVID-19. Neurosci. Behav. Physiol. 2021, 51, 856–866. [Google Scholar] [CrossRef]
- Lin, J.E.; Asfour, A.; Sewell, T.B.; Hooe, B.; Pryce, P.; Earley, C.; Shen, M.Y.; Kerner-Rossi, M.; Thakur, K.T.; Vargas, W.S.; et al. Neurological issues in children with COVID-19. Neurosci. Lett. 2020, 743, 135567. [Google Scholar] [CrossRef]
- Premraj, L.; Kannapadi, N.V.; Briggs, J.; Seal, S.M.; Battaglini, D.; Fanning, J.; Suen, J.; Robba, C.; Fraser, J.; Cho, S.-M. Mid and long-term neurological and neuropsychiatric manifestations of post-COVID-19 syndrome: A meta-analysis. J. Neurol. Sci. 2022, 434, 120162. [Google Scholar] [CrossRef]
- Rethinavel, H.S.; Ravichandran, S.; Radhakrishnan, R.K.; Kandasamy, M. COVID-19 and Parkinson’s disease: Defects in neurogenesis as the potential cause of olfactory system impairments and anosmia. J. Chem. Neuroanat. 2021, 115, 101965. [Google Scholar] [CrossRef]
- Barrett, M.J.; Murphy, J.M.; Zhang, J.; Blair, J.C.; Flanigan, J.L.; Nawaz, H.; Dalrymple, W.A.; Sperling, S.A.; Patrie, J.; Druzgal, T.J. Olfaction, cholinergic basal forebrain degeneration, and cognition in early Parkinson disease. Park. Relat. Disord. 2021, 90, 27–32. [Google Scholar] [CrossRef]
- Rahman, M.A.; Islam, K.; Rahman, S.; Alamin, M. Neurobiochemical Cross-talk Between COVID-19 and Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 1017–1023. [Google Scholar] [CrossRef]
- Rea, R.C.; Berlot, R.; Martin, S.L.; Craig, C.E.; Holmes, P.S.; Wright, D.J.; Bon, J.; Pirtošek, Z.; Ray, N.J. Quantitative EEG and cholinergic basal forebrain atrophy in Parkinson’s disease and mild cognitive impairment. Neurobiol. Aging 2021, 106, 37–44. [Google Scholar] [CrossRef]
- Courties, A.; Boussier, J.; Hadjadj, J.; Yatim, N.; Barnabei, L.; Péré, H.; Veyer, D.; Kernéis, S.; Carlier, N.; Pène, F.; et al. Regulation of the acetylcholine/α7nAChR anti-inflammatory pathway in COVID-19 patients. Sci. Rep. 2021, 11, 11886. [Google Scholar] [CrossRef]
- Kopańska, M.; Batoryna, M.; Bartman, P.; Szczygielski, J.; Banaś-Ząbczyk, A. Disorders of the Cholinergic System in COVID-19 Era—A Review of the Latest Research. Int. J. Mol. Sci. 2022, 23, 672. [Google Scholar] [CrossRef]
- Tizabi, Y.; Getachew, B.; Copeland, R.L.; Aschner, M. Nicotine and the nicotinic cholinergic system in COVID-19. FEBS J. 2020, 287, 3656–3663. [Google Scholar] [CrossRef]
- Schick, B.; Barth, E.; Mayer, B.; Weber, C.-L.; Hagemeyer, T.; Schönfeldt-Lecuona, C. Prospective, observational, single-centre cohort study with an independent control group matched for age and sex aimed at investigating the significance of cholinergic activity in patients with schizophrenia: Study protocol of the CLASH-study. BMJ Open 2021, 11, e050501. [Google Scholar] [CrossRef]
- Nakajima, K.; Abe, T.; Saji, R.; Ogawa, F.; Taniguchi, H.; Yamaguchi, K.; Sakai, K.; Nakagawa, T.; Matsumura, R.; Oi, Y.; et al. Serum cholinesterase associated with COVID-19 pneumonia severity and mortality. J. Infect. 2020, 82, 282–327. [Google Scholar] [CrossRef]
- Sejvar, J.J. Neurochemical and Neurobiological Weapons. Neurol. Clin. 2020, 38, 881–896. [Google Scholar] [CrossRef]
- Bekbossynova, A.; Zharylgap, A.; Filchakova, O. Venom-Derived Neurotoxins Targeting Nicotinic Acetylcholine Receptors. Molecules 2021, 26, 3373. [Google Scholar] [CrossRef]
- Caputi, V.; Giron, M. Microbiome-Gut-Brain Axis and Toll-Like Receptors in Parkinson’s Disease. Int. J. Mol. Sci. 2018, 19, 1689. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; O’Riordan, K.J.; Sandhu, K.; Peterson, V.; Dinan, T.G. The gut microbiome in neurological disorders. Lancet Neurol. 2020, 19, 179–194. [Google Scholar] [CrossRef]
- Sochocka, M.; Donskow-Łysoniewska, K.; Diniz, B.S.; Kurpas, D.; Brzozowska, E.; Leszek, J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease—A Critical Review. Mol. Neurobiol. 2019, 56, 1841–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popoff, M.R. Bacterial Toxins, Current Perspectives. Toxins 2020, 12, 570. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, A.; Travaglione, S.; Falzano, L.; Fiorentini, C. Bacterial Protein Toxins: Current and Potential Clinical Use. Curr. Med. Chem. 2008, 15, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Chiliveri, S.C.; Louis, J.M.; Ghirlando, R.; Bax, A. Transient lipid-bound states of spike protein heptad repeats provide insights into SARS-CoV-2 membrane fusion. Sci. Adv. 2021, 7, 2226. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Villaruz, A.E.; Li, M.; Cha, D.J.; Sturdevant, D.E.; Otto, M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 2006, 63, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.; Kong, B.; Moon, S.; Yu, S.-H.; Chung, J.; Ban, C.; Chung, W.-J.; Kim, S.-G.; Kweon, D.-H. Envelope-deforming antiviral peptide derived from influenza virus M2 protein. Biochem. Biophys. Res. Commun. 2019, 517, 507–512. [Google Scholar] [CrossRef]
- Solanki, S.S.; Singh, P.; Kashyap, P.; Sansi, M.S.; Ali, S.A. Promising role of defensins peptides as therapeutics to combat against viral infection. Microb. Pathog. 2021, 155, 104930. [Google Scholar] [CrossRef]
- Mustafa, S.; Balkhy, H.; Gabere, M. Peptide-Protein Interaction Studies of Antimicrobial Peptides Targeting Middle East Respiratory Syndrome Coronavirus Spike Protein: An In Silico Approach. Adv. Bioinform. 2019, 2019, 6815105. [Google Scholar] [CrossRef]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Querci, M.; Piazza, O.; Eede, G.V.D. Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients. F1000Research 2021, 10, 550. [Google Scholar] [CrossRef]
- Pistollato, F.; Petrillo, M.; Clerbaux, L.-A.; Leoni, G.; Ponti, J.; Bogni, A.; Brogna, C.; Cristoni, S.; Sanges, R.; Gyves, E.M.-D.; et al. Effects of spike protein and toxin-like peptides found in COVID-19 patients on human 3D neuronal/glial model undergoing differentiation: Possible implications for SARS-CoV-2 impact on brain development. Reprod. Toxicol. 2022, 111, 34–48. [Google Scholar] [CrossRef]
- Petrillo, M.; Brogna, C.; Cristoni, S.; Querci, M.; Piazza, O.; Eede, G.V.D. Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Research 2021, 10, 370. [Google Scholar] [CrossRef]
- Arzoni, A.; Bernardi, L.R.; Cristoni, S. In-source cloud ion mobility mass spectrometry. Rapid Commun. Mass Spectrom. 2015, 29, 690–694. [Google Scholar] [CrossRef]
- Cristoni, S.; Dusi, G.; Brambilla, P.; Albini, A.; Conti, M.; Brambilla, M.; Bruno, A.; Di Gaudio, F.; Ferlin, L.; Tazzari, V.; et al. SANIST: Optimization of a technology for compound identification based on the European Union directive with applications in forensic, pharmaceutical and food analyses. J. Mass Spectrom. 2017, 52, 16–21. [Google Scholar] [CrossRef]
- Cristoni, S.; Bernardi, L.R.; Larini, M.; Natale, G.; DiDomenico, N.; Varelli, M.; Conti, M.; Dorna, I.; Puccio, G. Predicting and preventing intestinal dysbiosis on the basis of pharmacological gut microbiota metabolism. Rapid Commun. Mass Spectrom. 2019, 33, 1221–1225. [Google Scholar] [CrossRef]
- Albini, A.; Briga, D.; Conti, M.; Bruno, A.; Farioli, D.; Canali, S.; Sogno, I.; D’Ambrosio, G.; Consonni, P.; Noonan, D.M. SANIST: A rapid mass spectrometric SACI/ESI data acquisition and elaboration platform for verifying potential candidate biomarkers. Rapid Commun. Mass Spectrom. 2015, 29, 1703–1710. [Google Scholar] [CrossRef] [Green Version]
- Madama, S.; Falletta, E.; Malvandi, A.M.; Arzoni, K.; Brogna, C.; Varelli, M.; Bertelli, M.; Conti, M.; Larini, M.; Guidugli, F.; et al. q value and parent energy optimization using a low voltage ionization approach increases resolution in linear ion trap mass spectrometry. J. Biol. Mass Spectrom. 2022, 57, e4876. [Google Scholar] [CrossRef]
- UniprotKB. Animal Toxin Annotation Project. Available online: https://www.uniprot.org/program/Toxins (accessed on 4 October 2020).
- Arike, L.; Peil, L. Spectral Counting Label-Free Proteomics. Methods Mol. Biol. 2014, 1156, 213–222. [Google Scholar]
- Lundgren, D.H.; Hwang, S.; Wu, L.; Han, D.K. Role of spectral counting in quantitative proteomics. Expert Rev. Proteom. 2010, 7, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, K.H. SARS-CoV-2 spike glycoprotein-binding proteins expressed by upper respiratory tract bacteria may prevent severe viral infection. FEBS Lett. 2020, 594, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Dragelj, J.; Mroginski, M.A.; Ebrahimi, K.H. Hidden in Plain Sight: Natural Products of Commensal Microbiota as an Environmental Selection Pressure for the Rise of New Variants of SARS-CoV-2. Chembiochem 2021, 22, 2946–2950. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-S.; Kao, C.-Y. Current understanding of the gut microbiota shaping mechanisms. J. Biomed. Sci. 2019, 26, 59. [Google Scholar] [CrossRef] [PubMed]
- Greig, N.H.; Reale, M.; Tata, A.M. New Pharmacological Approaches to the Cholinergic System: An Overview on Muscarinic Receptor Ligands and Cholinesterase Inhibitors. Recent Pat. CNS Drug Discov. 2013, 8, 123–141. [Google Scholar] [CrossRef]
- Le Jeune, H.; Aubert, I.; Jourdan, F.; Quirion, R. Comparative laminar distribution of various autoradiographic cholinergic markers in adult rat main olfactory bulb. J. Chem. Neuroanat. 1995, 9, 99–112. [Google Scholar] [CrossRef]
- Alkondon, M.; Rocha, E.S.; Maelicke, A.; Albuquerque, E.X. Diversity of nicotinic acetylcholine receptors in rat brain. V. alpha-Bungarotoxin-sensitive nicotinic receptors in olfactory bulb neurons and presynaptic modulation of glutamate release. J. Pharmacol. Exp. Ther. 1996, 278, 1460–1471. [Google Scholar]
- Bucaretchi, F.; Borrasca-Fernandes, C.F.; De Capitani, E.M.; Hyslop, S. Consecutive envenomation of two men bitten by the same coral snake (Micrurus corallinus). Clin. Toxicol. 2019, 58, 132–135. [Google Scholar] [CrossRef]
- Sethi, M.; Cook, M.; Winkel, K.D. Persistent anosmia and olfactory bulb atrophy after mulga (Pseudechis australis) snakebite. J. Clin. Neurosci. 2016, 29, 199–201. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, nrdp201763, Erratum in Nat. Rev. Dis. Prim. 2017, 3, 17079. [Google Scholar] [CrossRef]
- Pearn, J.; McGuire, B.; McGuire, L.; Richardson, P. The envenomation syndrome caused by the Australian Red-bellied Black Snake Pseudechis porphyriacus. Toxicon 2000, 38, 1715–1729. [Google Scholar] [CrossRef]
- Dennis, E.A.; Cao, J.; Hsu, Y.-H.; Magrioti, V.; Kokotos, G. Phospholipase A2 Enzymes: Physical Structure, Biological Function, Disease Implication, Chemical Inhibition, and Therapeutic Intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Sig. Transduct. Target. Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Ragheb, F.; Molina-Holgado, E.; Cui, Q.-L.; Khorchid, A.; Liu, H.-N.; LaRocca, J.N.; Almazan, G. Pharmacological and functional characterization of muscarinic receptor subtypes in developing oligodendrocytes. J. Neurochem. 2001, 77, 1396–1406. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, F.; Bernardo, A.; Magnaghi, V.; Minghetti, L.; Tata, A.M. Muscarinic receptor subtypes as potential targets to modulate oligodendrocyte progenitor survival, proliferation, and differentiation. Dev. Neurobiol. 2012, 72, 713–728. [Google Scholar] [CrossRef]
- Brogna, C.; Brogna, B.; Bisaccia, D.R.; Lauritano, F.; Marino, G.; Montano, L.; Cristoni, S.; Prisco, M.; Piscopo, M. Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages? Vaccines 2022, 10, 708. [Google Scholar] [CrossRef]
- Carvelli, J.; Demaria, O.; Vély, F.; Batista, L.; Benmansour, N.C.; Fares, J.; Carpentier, S.; Thibult, M.L.; Morel, A.; André, P.; et al. Gut Microbiota from Patients with Mild COVID-19 Cause Alterations in Mice that Resemble Post-COVID Syndrome (Version 1). Research Square 2022. [Google Scholar] [CrossRef]
- Moss, B.; Rosenblum, E.N.; Katz, E.; Grimley, P.M. Rifampicin: A Specific Inhibitor of Vaccinia Virus Assembly. Nature 1969, 224, 1280–1284. [Google Scholar] [CrossRef]
- Fujinaga, Y.; Kawaratani, H.; Kaya, D.; Tsuji, Y.; Ozutsumi, T.; Furukawa, M.; Kitagawa, K.; Sato, S.; Nishimura, N.; Sawada, Y.; et al. Effective Combination Therapy of Angiotensin-II Receptor Blocker and Rifaximin for Hepatic Fibrosis in Rat Model of Nonalcoholic Steatohepatitis. Int. J. Mol. Sci. 2020, 21, 5589. [Google Scholar] [CrossRef]
- Shayto, R.H.; Mrad, R.A.; Sharara, A.I. Use of rifaximin in gastrointestinal and liver diseases. World J. Gastroenterol. 2016, 22, 6638–6651. [Google Scholar] [CrossRef] [PubMed]
- Llor, C.; Pérez, A.; Carandell, E.; García-Sangenís, A.; Rezola, J.; Llorente, M.; Gestoso, S.; Bobé, F.; Román-Rodríguez, M.; Cots, J.M.; et al. Efficacy of high doses of penicillin versus amoxicillin in the treatment of uncomplicated community acquired pneumonia in adults. A non-inferiority controlled clinical trial. Aten Primaria 2019, 51, 32–39. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, P.W.; Claesson, M.J. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int. Dairy J. 2010, 20, 281–291. [Google Scholar] [CrossRef]
- Rondanelli, M. Review on microbiota and effectiveness of probiotics use in older. World J. Clin. Cases 2015, 3, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Mäkivuokko, H.; Tiihonen, K.; Tynkkynen, S.; Paulin, L.; Rautonen, N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br. J. Nutr. 2009, 103, 227–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cyprian, F.; Sohail, M.U.; Abdelhafez, I.; Salman, S.; Attique, Z.; Kamareddine, L.; Al-Asmakh, M. SARS-CoV-2 and immune-microbiome interactions: Lessons from respiratory viral infections. Int. J. Infect. Dis. 2021, 105, 540–550. [Google Scholar] [CrossRef]
- Brogna, B.; Brogna, C.; Petrillo, M.; Conte, A.M.; Benincasa, G.; Montano, L.; Piscopo, M. SARS-CoV-2 Detection in Fecal Sample from a Patient with Typical Findings of COVID-19 Pneumonia on CT but Negative to Multiple SARS-CoV-2 RT-PCR Tests on Oropharyngeal and Nasopharyngeal Swab Samples. Medicina 2021, 57, 290. [Google Scholar] [CrossRef]
- Pang, I.K.; Iwasaki, A. Control of antiviral immunity by pattern recognition and the microbiome. Immunol. Rev. 2012, 245, 209–226. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Dakshinamoorthy, M.; Venkatesh, A.; Arumugam, K. A literature review on dental caries vaccine-A prevention strategy. Indian J. Public Health Res. Dev. 2019, 10, 3041–3043. [Google Scholar]



| Drugs | Viral RNA Load | Toxins Aspect |
|---|---|---|
| Rifaximin | Decrease - | Not present |
| Azithromycin | Decrease ---- | Present + |
| Erythromycin | Increase + | Present ++ |
| Metronidazole | Decrease ---- | Present ++ |
| Clindamycin | Not change | Present +++ |
| Lincomycin | Increase + | Present +++ |
| Piperacillin + tazobactam | Decrease -- | Present + |
| Vancomycin | Decrease ---- | Present + |
| Amoxicillin | Decrease ---- | Present + |
| Ampicillin | Decrease -- | Present + |
| Cefixime | Decrease --- | Present + |
| Ceftriaxone | Decrease -- | Present + |
| Meropenem | Decrease - | Present ++ |
| Gentamicin | Decrease - | Present ++ |
| Ciprofloxacin | Decrease -- | Present ++ |
| Colistin | Increase + | Present ++ |
| Teicoplanin | Decrease -- | Present + |
| Levofloxacin | Increase ++ | Present ++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brogna, C.; Cristoni, S.; Brogna, B.; Bisaccia, D.R.; Marino, G.; Viduto, V.; Montano, L.; Piscopo, M. Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2—New Data for a Possible Role in the Long COVID Pattern. Biomedicines 2023, 11, 87. https://doi.org/10.3390/biomedicines11010087
Brogna C, Cristoni S, Brogna B, Bisaccia DR, Marino G, Viduto V, Montano L, Piscopo M. Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2—New Data for a Possible Role in the Long COVID Pattern. Biomedicines. 2023; 11(1):87. https://doi.org/10.3390/biomedicines11010087
Chicago/Turabian StyleBrogna, Carlo, Simone Cristoni, Barbara Brogna, Domenico Rocco Bisaccia, Giuliano Marino, Valentina Viduto, Luigi Montano, and Marina Piscopo. 2023. "Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2—New Data for a Possible Role in the Long COVID Pattern" Biomedicines 11, no. 1: 87. https://doi.org/10.3390/biomedicines11010087
APA StyleBrogna, C., Cristoni, S., Brogna, B., Bisaccia, D. R., Marino, G., Viduto, V., Montano, L., & Piscopo, M. (2023). Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2—New Data for a Possible Role in the Long COVID Pattern. Biomedicines, 11(1), 87. https://doi.org/10.3390/biomedicines11010087